
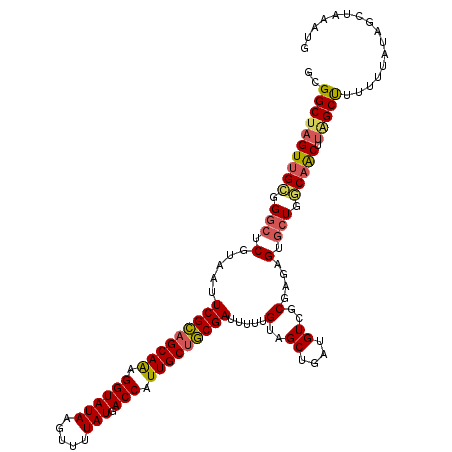
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,617,487 – 11,617,604 |
| Length | 117 |
| Max. P | 0.975090 |

| Location | 11,617,487 – 11,617,604 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -33.82 |
| Energy contribution | -33.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
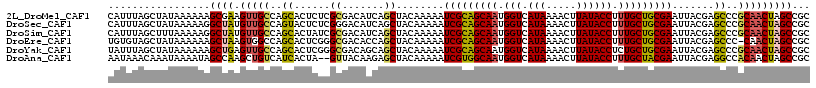
>2L_DroMel_CAF1 11617487 117 + 22407834 GCGGCUAGUUGCGGGCUCGUAAUUCGCAGCAAAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGAUGUCGCGAGAGUGCUGGCAACUUCGCUUUUUUAUAGCUAAAUG ((((..((((((.(((.(.....(((((((((.((((((.....))).))).))))))))).(((((..((....))..)))))).))).))))))))))................. ( -43.10) >DroSec_CAF1 70788 117 + 1 GCGGCUAGUUGCGGGCUCGUAAUUCGCAGCAAAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGAUGUCCCGAGAGUACUGGCAACAUAGCCUUUUUAUAGCUAAAUG ..((((((((((.(((((.....(((((((((.((((((.....))).))).)))))))))...(((..((....))..))))))..)).))))).)))))................ ( -40.40) >DroSim_CAF1 74574 117 + 1 GCGGCUAGUUGCGGGCUCGUAAUUCGCAGCAAAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGAUGUCGCGAUAGUGCUGGCAACAUAGCCUUUUUAAAGCUAAAUG ..((((((((((.(((.(.....(((((((((.((((((.....))).))).)))))))))...(((..((....))..)))..).))).))))).)))))................ ( -44.10) >DroEre_CAF1 101259 116 + 1 GCGGCUAGUUG-GGGCUCGUAAUUCGCAGCAAAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGGUGUCGCCCGAGUGCUGGCCACUUAGCUUUUUUAUAGCUACACA ..((((((((.-((((.......(((((((((.((((((.....))).))).)))))))))........((....)).)))).)..)))))))...(((((.......))))).... ( -41.40) >DroYak_CAF1 74001 117 + 1 GCGGCUAGUUGCGGGCUCGUAAUUCGCAGCAGAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGCUGUCGCCCGAGUGCUGGCAACUCAGCUUUUUUAUAGCUAAAUA (..((((((..(((((.......(((((((((.((((((.....))).))).)))))))))........((....)).)))))...))))))..)..((((.......))))..... ( -40.70) >DroAna_CAF1 83965 115 + 1 GCGGCUAGUUGUGGCCUCGUAAUUCGUAGCAAAGGUAUAAGUUUUAUGACCAUUGCCACGAUUUUUGUAGCUCUUGUAAC--UAGUGAUGACAGCUUGGCUAUUUUAUUUGUUUAUU ..(((((((((((((..((.((.((((.((((.((((((.....))).))).)))).)))).)).))..)))........--((....))))))).)))))................ ( -25.40) >consensus GCGGCUAGUUGCGGGCUCGUAAUUCGCAGCAAAGGUAUAAGUUUUAUGACCAUUGCUGCGAUUUUUGUAGCUGAUGUCGCGAGAGUGCUGGCAACUUAGCUUUUUUAUAGCUAAAUG ..((((((((((.(((.(.....(((((((((.((((((.....))).))).))))))))).....(..((....))..)....).))).))))).)))))................ (-33.82 = -33.88 + 0.06)


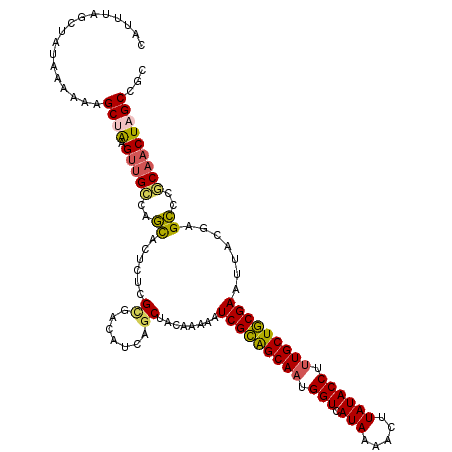
| Location | 11,617,487 – 11,617,604 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -27.45 |
| Energy contribution | -27.32 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
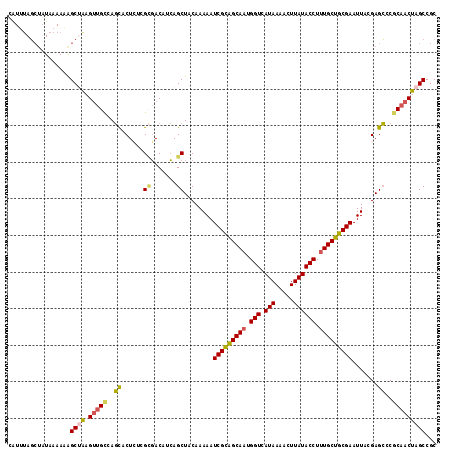
>2L_DroMel_CAF1 11617487 117 - 22407834 CAUUUAGCUAUAAAAAAGCGAAGUUGCCAGCACUCUCGCGACAUCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUUUGCUGCGAAUUACGAGCCCGCAACUAGCCGC .................((..((((((..((......((.......))........(((((((((.(((.(((.....)))))).))))))))).......))..)))))).))... ( -32.20) >DroSec_CAF1 70788 117 - 1 CAUUUAGCUAUAAAAAGGCUAUGUUGCCAGUACUCUCGGGACAUCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUUUGCUGCGAAUUACGAGCCCGCAACUAGCCGC ................(((((.(((((..((.((....))))....(((.......(((((((((.(((.(((.....)))))).)))))))))......)))..)))))))))).. ( -37.02) >DroSim_CAF1 74574 117 - 1 CAUUUAGCUUUAAAAAGGCUAUGUUGCCAGCACUAUCGCGACAUCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUUUGCUGCGAAUUACGAGCCCGCAACUAGCCGC ................(((((.(((((..((......((.......))........(((((((((.(((.(((.....)))))).))))))))).......))..)))))))))).. ( -37.80) >DroEre_CAF1 101259 116 - 1 UGUGUAGCUAUAAAAAAGCUAAGUGGCCAGCACUCGGGCGACACCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUUUGCUGCGAAUUACGAGCCC-CAACUAGCCGC ....(((((.......))))).(((((.((..(((((((.......))).......(((((((((.(((.(((.....)))))).)))))))))....))))...-...)).))))) ( -36.40) >DroYak_CAF1 74001 117 - 1 UAUUUAGCUAUAAAAAAGCUGAGUUGCCAGCACUCGGGCGACAGCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUCUGCUGCGAAUUACGAGCCCGCAACUAGCCGC ...((((((.......))))))((((((........)))))).((.(((.......((((((((..(((.(((.....))))))..))))))))......)))..)).......... ( -36.82) >DroAna_CAF1 83965 115 - 1 AAUAAACAAAUAAAAUAGCCAAGCUGUCAUCACUA--GUUACAAGAGCUACAAAAAUCGUGGCAAUGGUCAUAAAACUUAUACCUUUGCUACGAAUUACGAGGCCACAACUAGCCGC ..............(((((...)))))..((..((--(((.....)))))......(((((((((.(((.(((.....)))))).))))))))).....))(((........))).. ( -24.20) >consensus CAUUUAGCUAUAAAAAAGCUAAGUUGCCAGCACUCUCGCGACAUCAGCUACAAAAAUCGCAGCAAUGGUCAUAAAACUUAUACCUUUGCUGCGAAUUACGAGCCCGCAACUAGCCGC .................((((.(((((..((......((.......))........(((((((((.(((.(((.....)))))).))))))))).......))..)))))))))... (-27.45 = -27.32 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:37 2006