| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,217,355 – 1,217,449 |
| Length | 94 |
| Max. P | 0.662560 |

| Location | 1,217,355 – 1,217,449 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
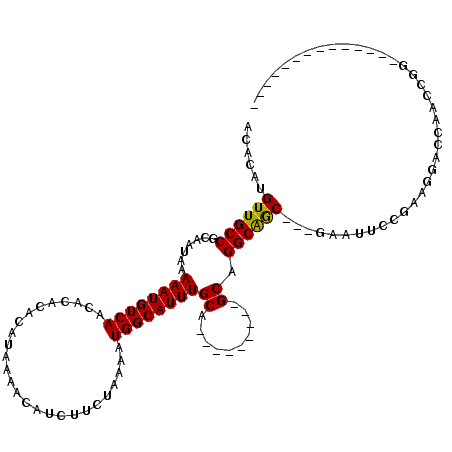
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -13.93 |
| Energy contribution | -13.57 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
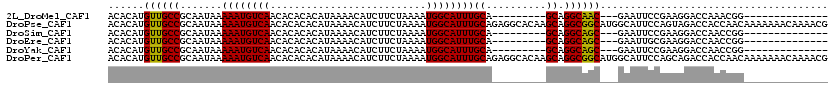
>2L_DroMel_CAF1 1217355 94 - 22407834 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCA---------GCAGGCAAC---GAAUUCCGAAGGACCAAACGG-------------- .....(((((((((.....((((((((..........................))))))))...---------)).))))))---)....(((..........)))-------------- ( -17.27) >DroPse_CAF1 16861 120 - 1 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCAGAGGCACAAGCAGGCGGCAUGGCAUUCCAGUAGACCACCAACAAAAAAACAAAACG ...(((((((((((.....((((((((..........................))))))))((....))....)).)))))))))................................... ( -22.77) >DroSim_CAF1 13004 94 - 1 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCA---------GCAGGCAGC---GAAUUCCGAAGGACCAACCGG-------------- .....(((((((((.....((((((((..........................))))))))...---------)).))))))---)....(((..........)))-------------- ( -17.67) >DroEre_CAF1 12731 94 - 1 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCA---------GCAGGCAGC---GAAUUGCGAAGGACCAACCGG-------------- .....(((((((((.....((((((((..........................))))))))...---------)).))))))---).........((.....))..-------------- ( -17.57) >DroYak_CAF1 13796 94 - 1 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCA---------GCAGGCAGC---GAAUUCCGAAGGACCAACCGG-------------- .....(((((((((.....((((((((..........................))))))))...---------)).))))))---)....(((..........)))-------------- ( -17.67) >DroPer_CAF1 16665 120 - 1 ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCAGAGGCACAAGCAGGCGGCAUGGCAUUCCAGCAGACCACCAACAAAAAAACAAAACG ...(((((((((((.....((((((((..........................))))))))((....))....)).)))))))))((......))......................... ( -23.97) >consensus ACACAUGUUGCCGCAAUAAAAAUGUCAACACACACAUAAAACAUCUUCUAAAAUGGCAUUUGCA_________GCAGGCAGC___GAAUUCCGAAGGACCAACCGG______________ ......((((((.......((((((((..........................))))))))((..........)).))))))...................................... (-13.93 = -13.57 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:59 2006