
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,609,330 – 11,609,422 |
| Length | 92 |
| Max. P | 0.999053 |

| Location | 11,609,330 – 11,609,422 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -19.34 |
| Energy contribution | -18.59 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.27 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11609330 92 + 22407834 UAAGAAAGAGCGCAAAAAGCGCAAAA--CGCAAAGUGAAGAGCUAUUCUACAUACAUG----------UAUAUAUGUAUGUAUGUGUGGGUGGAUGCUUGCAGA .........((((.....))))....--...........((((...(((((.((((((----------(((((....))))))))))).))))).))))..... ( -26.40) >DroSec_CAF1 63949 101 + 1 UAAGAAAGAGCGCAAAAAGCGCAAAA--CGCAAAGUGAAGAGCUAU-CUACAUACAUGAACGUACAUACAUACGUAUAUGUAUGUAUGGGUGGAUGCUUGCAGA .........((((.....))))....--.((((.(((.....((((-(((((((((((.(((((......))))).)))))))))..)))))).)))))))... ( -30.80) >DroSim_CAF1 66725 101 + 1 UAAGAAAGAGCGCAAAAAGCGCAAAA--CGCAAAGUGAAGAGCUAU-CUACAUACAUGAACCUACAUACAUAUAUGUAUGUAUAUAUGGGUGGAUGCUUGCAGA .........((((.....))))....--...........((((.((-((((...((((....((((((((....))))))))..)))).))))))))))..... ( -27.70) >DroEre_CAF1 94592 97 + 1 UAAGAAAGAGCGCCAAAUGCGCAAAAAGCGCAAAGUGAAGAGCUAU-CCGCAUACAUGGAUGUAUGUG--UACGUGUGCGUGUG----UGUGGGUGGUUGCAGA .........((..((..(((((.....)))))...))...((((((-(((((((((((.(((((....--)))))...))))))----)))))))))))))... ( -36.90) >consensus UAAGAAAGAGCGCAAAAAGCGCAAAA__CGCAAAGUGAAGAGCUAU_CUACAUACAUGAACGUACAUACAUACAUGUAUGUAUGUAUGGGUGGAUGCUUGCAGA .........((((.....)))).......(((((((.....)))....((((((((((..............))))))))))...............))))... (-19.34 = -18.59 + -0.75)

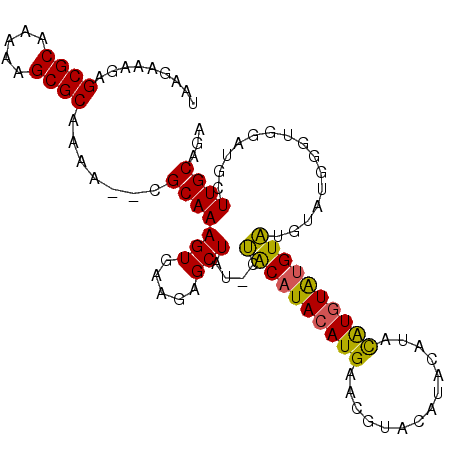
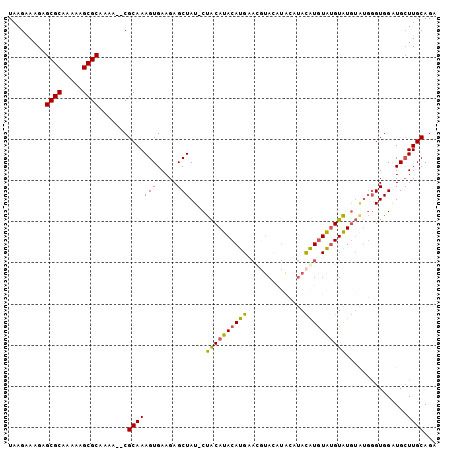
| Location | 11,609,330 – 11,609,422 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11609330 92 - 22407834 UCUGCAAGCAUCCACCCACACAUACAUACAUAUAUA----------CAUGUAUGUAGAAUAGCUCUUCACUUUGCG--UUUUGCGCUUUUUGCGCUCUUUCUUA ...(((((..............(((((((((.....----------.)))))))))(((......)))..))))).--....((((.....))))......... ( -20.00) >DroSec_CAF1 63949 101 - 1 UCUGCAAGCAUCCACCCAUACAUACAUAUACGUAUGUAUGUACGUUCAUGUAUGUAG-AUAGCUCUUCACUUUGCG--UUUUGCGCUUUUUGCGCUCUUUCUUA ((((((.((((..((.(((((((((......)))))))))...))..)))).)))))-)..((..........)).--....((((.....))))......... ( -28.00) >DroSim_CAF1 66725 101 - 1 UCUGCAAGCAUCCACCCAUAUAUACAUACAUAUAUGUAUGUAGGUUCAUGUAUGUAG-AUAGCUCUUCACUUUGCG--UUUUGCGCUUUUUGCGCUCUUUCUUA ((((((.((((..(((..((((((((((....)))))))))))))..)))).)))))-)..((..........)).--....((((.....))))......... ( -28.50) >DroEre_CAF1 94592 97 - 1 UCUGCAACCACCCACA----CACACGCACACGUA--CACAUACAUCCAUGUAUGCGG-AUAGCUCUUCACUUUGCGCUUUUUGCGCAUUUGGCGCUCUUUCUUA (((((...........----...(((....))).--...(((((....)))))))))-).(((.((......(((((.....)))))...)).)))........ ( -20.20) >consensus UCUGCAAGCAUCCACCCAUACAUACAUACACAUAUGUAUGUACGUUCAUGUAUGUAG_AUAGCUCUUCACUUUGCG__UUUUGCGCUUUUUGCGCUCUUUCUUA ...(((((..............((((((((((..............))))))))))..............))))).......((((.....))))......... (-14.90 = -15.90 + 1.00)

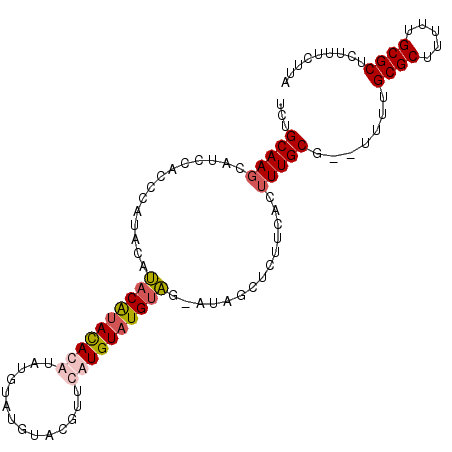
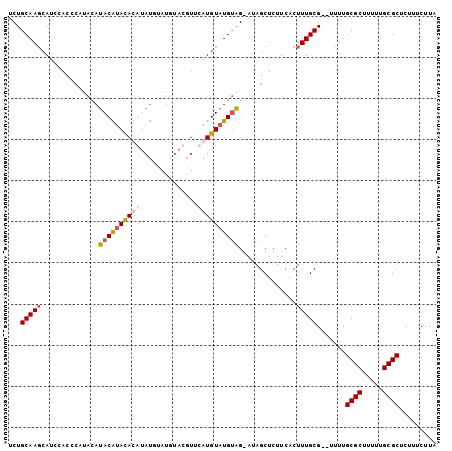
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:32 2006