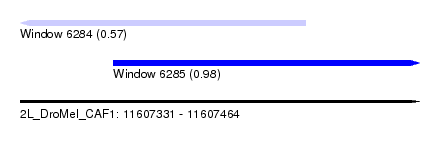
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,607,331 – 11,607,464 |
| Length | 133 |
| Max. P | 0.981511 |

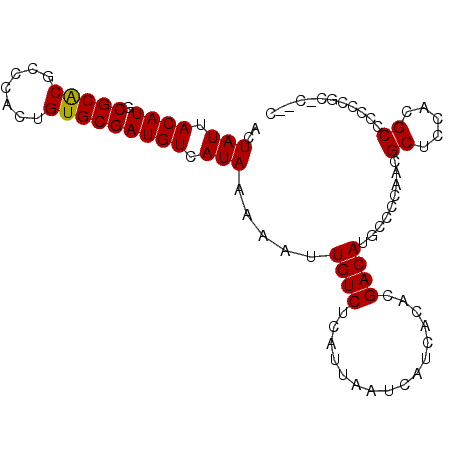
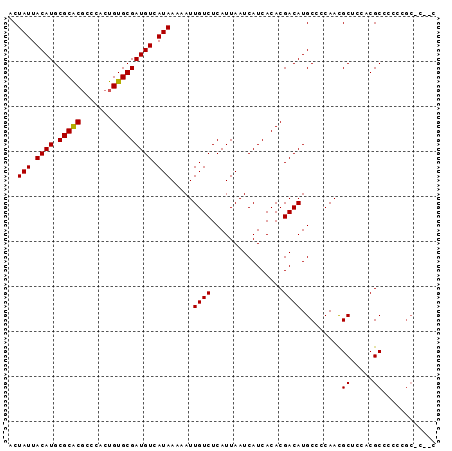
| Location | 11,607,331 – 11,607,426 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -12.96 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.29 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11607331 95 - 22407834 ACUAUUACAUGCGCACGCCCACUGUGCGAUGUCAUAAAAAUUGUCUCAUUAAUCAUCACACGACAUGCCCCAACGCUCCACGCUCCACGCUCCAC ..(((.((((.((((((.....)))))))))).))).....((((................)))).((......)).....((.....))..... ( -13.39) >DroSec_CAF1 61907 95 - 1 ACUAUUACAUGCGCACGCCCACUGUGCGAUGUCAUAAAAAUUGUCUCAUUAAUCAUCACACGACAUGCCCCAAUGCUCCACGCCCUCCGCCCACC ..(((.((((.((((((.....)))))))))).))).....((((................)))).((......((.....)).....))..... ( -12.89) >DroEre_CAF1 92790 88 - 1 ACUAUUACAUGCGCGCUCCCACUGUGCGAUGUCAUAAAAAUUGUCUCAUUAAUCAUCACACGACAUGCCCCAACGCUCCACGCCCCC-------U ..(((.((((.(((((.......))))))))).))).....((((................)))).........((.....))....-------. ( -12.59) >consensus ACUAUUACAUGCGCACGCCCACUGUGCGAUGUCAUAAAAAUUGUCUCAUUAAUCAUCACACGACAUGCCCCAACGCUCCACGCCCCCCGC_C__C ..(((.((((.(((((.......))))))))).))).....((((................)))).........((.....))............ (-12.51 = -12.29 + -0.22)

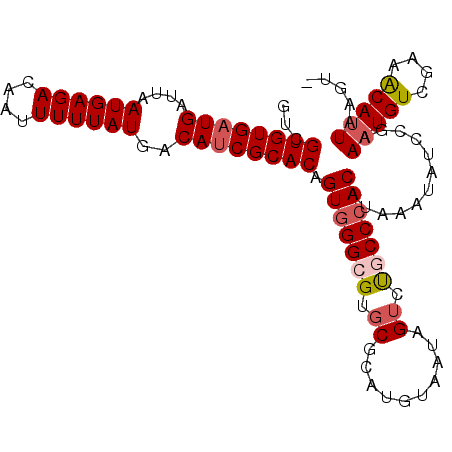
| Location | 11,607,362 – 11,607,464 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11607362 102 + 22407834 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGCGUGCGCAUGUAAUAGUCUGCCCACAUAAAUAUCCGAAAUGUCGAAACAUUAAGU- .((((((((((....((((((....))))))..)))))))).(((((((.((..........)).)))))))..........))(((((....)))))....- ( -28.90) >DroSec_CAF1 61938 102 + 1 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGCGUGCGCAUGUAAUAGUCUGCCCACAUAAAUAUCCGAAAUGUCGAAACAUUAAGU- .((((((((((....((((((....))))))..)))))))).(((((((.((..........)).)))))))..........))(((((....)))))....- ( -28.90) >DroSim_CAF1 64702 102 + 1 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAGCGCACAGUGGGCGUGCGUAUGUAAUAGUCUGCCCACAUAAAUAUCCGAAAUGUCGAAACAUUAAGU- .(((((((.((....((((((....))))))..)).))))).(((((((.((..........)).)))))))..........))(((((....)))))....- ( -24.30) >DroEre_CAF1 92814 102 + 1 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGAGCGCGCAUGUAAUAGUCUGCCCACAUAAAUAUCCAAAAUGUCGAAACAUUAAGU- ...((((((((....((((((....))))))..)))))))).(((((((.((..........)))).)))))............(((((....)))))....- ( -23.90) >DroYak_CAF1 64030 102 + 1 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGAGAGCGCAUGUAAUAGUCUGCCCACAUAAAUAUCCGAAAUGUCGAAACACUAAGU- ...((((((((....((((((....))))))..)))))))).((((((((.(..........)))).))))).........(((....)))...........- ( -24.00) >DroAna_CAF1 75973 95 + 1 GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGCGA----AUGUC----UACGCCAACACAAAUAUCAGAAAUGUCGAGGCAUUAAGAA .((((((((((....((((((....))))))..)))))))).(((((((.----.....----..))))..)))........))(((((....)))))..... ( -24.50) >consensus GUCGUGUGAUGAUUAAUGAGACAAUUUUUAUGACAUCGCACAGUGGGCGUGCGCAUGUAAUAGUCUGCCCACAUAAAUAUCCGAAAUGUCGAAACAUUAAGU_ ...((((((((....((((((....))))))..)))))))).(((((((.((..........)).)))))))............(((((....)))))..... (-24.20 = -24.97 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:29 2006