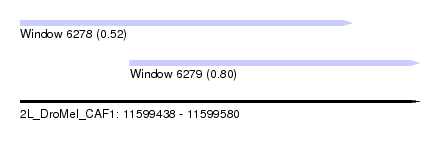
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,599,438 – 11,599,580 |
| Length | 142 |
| Max. P | 0.803641 |

| Location | 11,599,438 – 11,599,556 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11599438 118 + 22407834 GCGAUGGCAACGCCCACU-UUAUUUGCUACAAAACAAUUUCCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCA ....((((((((((((..-....((((((((.........................((((......))))(((((((....)).))))-)...))))))))..))))).....))))))) ( -34.00) >DroPse_CAF1 51429 119 + 1 GUCAUGCCAACGCCCAUUUUUAUUUGCUACAAAACAAUUUUCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUACACAA-AAUGUGUGGCAAUUUCGACAAACAGUUGCCA ....((.((((............((((((((............((((.(((....(((((......))))).))).))))........-....))))))))............)))).)) ( -20.70) >DroSim_CAF1 56287 118 + 1 GCGAUAGCAACCCCCUUU-UUAUUUGCUACAAAACAAUUUCCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCA ((....))..........-....((((((((.........................((((......))))(((((((....)).))))-)...))))))))....(((((....))))). ( -25.20) >DroYak_CAF1 55295 118 + 1 GCGAUGGCAACGCCCAUU-UUAUUUGCUACAAAACAAUUUCCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCA ....((((((((((((..-....((((((((.........................((((......))))(((((((....)).))))-)...))))))))..))))).....))))))) ( -34.70) >DroAna_CAF1 67147 118 + 1 GCAAUGGCUACGCCCAUU-UUAUUUCCUACAAAACAAUUUCGAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCGCAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCA (((((......(((((..-((.((((...............))))(((((((.((.((((((.((.(((....))).)).))).))).-)).)))))))))..))))).....))))).. ( -28.76) >DroPer_CAF1 53792 120 + 1 GUCAUGCCAACGCCCAUUUUUAUUUGCUACAAAACAAUUUUCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUACACAAAAAUGUGUGGCAAUUUCGACAAACAGUUGCCA ....((.((((............((((((((........................(((((......)))))....(((((........)))))))))))))............)))).)) ( -20.45) >consensus GCGAUGGCAACGCCCAUU_UUAUUUGCUACAAAACAAUUUCCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA_AAUGUGUGGCAAUUUGGGCAAACAGUUGCCA ....(((((((............((((((((............((((.(((....(((((......))))).))).)))).............))))))))............))))))) (-21.69 = -22.53 + 0.83)

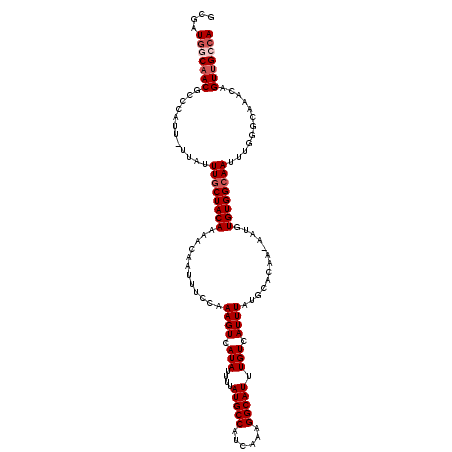
| Location | 11,599,477 – 11,599,580 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -18.09 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11599477 103 + 22407834 CCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCAAAACAGA--------------ACUAUGUAGUUU--AGAGC .(((((((((((....((((......))))(((((((....)).))))-)..)))))))..))))(((((....)))))......((--------------(((....)))))--..... ( -22.70) >DroPse_CAF1 51469 88 + 1 UCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUACACAA-AAUGUGUGGCAAUUUCGACAAACAGUUGCCACAAGGAA--------------AC----------------- ...((((.(((....(((((......))))).))).))))........-....((((((((((.........)))))))))).....--------------..----------------- ( -19.10) >DroEre_CAF1 84582 103 + 1 CCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCAAAACAGA--------------ACUGUGUAGUUU--AGAGC ...((((.(((....(((((......))))).))).)))).((((((.-..(((.((((((((.(......)))))))))..)))..--------------..))))))....--..... ( -24.50) >DroYak_CAF1 55334 103 + 1 CCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCAAAACAGA--------------ACUAUGUAGUUU--GGAGC (((((.(((((.....((((......))))(((((.......((((..-...))))(((((((.(......))))))))...)))))--------------..)))).).)))--))... ( -26.90) >DroAna_CAF1 67186 119 + 1 CGAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCGCAA-AAUGUGUGGCAAUUUGGGCAAACAGUUGCCAAAACAGAGCCUUACCAGGAAAACGUUGUAGUUCAUGGGAC .....(((................(((((.(((((.......((((..-...))))(((((((.(......))))))))...)))))))))).(((....(((......)))..)))))) ( -27.40) >DroPer_CAF1 53832 88 + 1 UCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUACACAAAAAUGUGUGGCAAUUUCGACAAACAGUUGCCACAAGGA---------------AC----------------- ...((((.(((....(((((......))))).))).)))).............((((((((((.........))))))))))....---------------..----------------- ( -19.10) >consensus CCAAAGUCAUAUUUUAUGCCAUCAAAGGCAUUUGUCAUUUAUGCACAA_AAUGUGUGGCAAUUUGGGCAAACAGUUGCCAAAACAGA______________ACU_UGUAGUUU___GAGC ...((((.(((....(((((......))))).))).))))..(((((....)))))(((((((.........)))))))......................................... (-18.09 = -17.73 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:23 2006