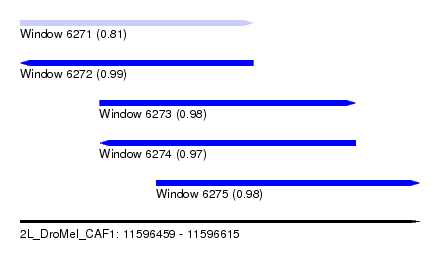
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,596,459 – 11,596,615 |
| Length | 156 |
| Max. P | 0.987785 |

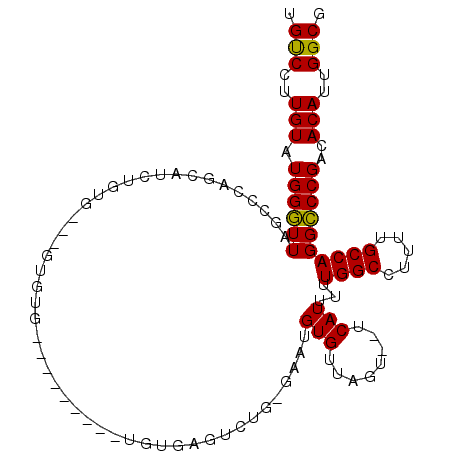
| Location | 11,596,459 – 11,596,550 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.03 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11596459 91 + 22407834 UGUCCUUGUAUGGGUUAGCCCAGCAUCUGUG-----------------UGUGAGUCUGCGAAUGUGUUAGU--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCG ..........((((....))))((....(((-----------------((((.(((((((((.(.(((((.--......)))))).)))).))))).).))))))..)). ( -26.90) >DroYak_CAF1 52581 101 + 1 UGUCCUUGUAUGGGUUAGUCCAGCAUCUGUGUGUGUGUGAGC-------ACAUCCUUGUGGAUGUGUUAGU--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCG ..........((((....))))((....(((((((((..(((-------((((((....)))))))))...--.)))....(((((......)))))..))))))..)). ( -33.40) >DroAna_CAF1 64548 100 + 1 -GCCAGUGUAUGGCUUGGCUUAGA----GUGCACGUGUG---UGUGUGUGUGAGUGGC--UGUGUGUUAGUGCUCAUUUUUGGCCUUUUGCCAGGGCCGACACAUUGGCG -((((((((.(((((((((..((.----(((((((....---..)))))(((((((.(--((.....)))))))))).....))))...)))).)))))..)))))))). ( -38.30) >consensus UGUCCUUGUAUGGGUUAGCCCAGCAUCUGUG___GUGUG_________UGUGAGUCUG_GAAUGUGUUAGU__UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCG .(((..(((.((((((...............................................(((........)))...((((.....))))))))))..)))..))). (-17.03 = -16.37 + -0.66)

| Location | 11,596,459 – 11,596,550 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.03 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -11.81 |
| Energy contribution | -11.37 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11596459 91 - 22407834 CGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA--ACUAACACAUUCGCAGACUCACA-----------------CACAGAUGCUGGGCUAACCCAUACAAGGACA .((.(((((((..(((((......)))))........--....))))))).)).........-----------------.........((((....)))).......... ( -21.94) >DroYak_CAF1 52581 101 - 1 CGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA--ACUAACACAUCCACAAGGAUGU-------GCUCACACACACACAGAUGCUGGACUAACCCAUACAAGGACA .((...(((((.((((((......)))))........--.....(((((((....))))))-------)........))))))...))(((......))).......... ( -28.90) >DroAna_CAF1 64548 100 - 1 CGCCAAUGUGUCGGCCCUGGCAAAAGGCCAAAAAUGAGCACUAACACACA--GCCACUCACACACACA---CACACGUGCAC----UCUAAGCCAAGCCAUACACUGGC- .((((.(((((.(((..((((....(((......................--))).........(((.---.....)))...----.....)))).)))))))).))))- ( -26.25) >consensus CGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA__ACUAACACAUAC_CAAACUCACA_________CACAC___CACAGAUGCUGGGCUAACCCAUACAAGGACA ......(((((.(((..((((.....))))....((........))..................................................))))))))...... (-11.81 = -11.37 + -0.44)

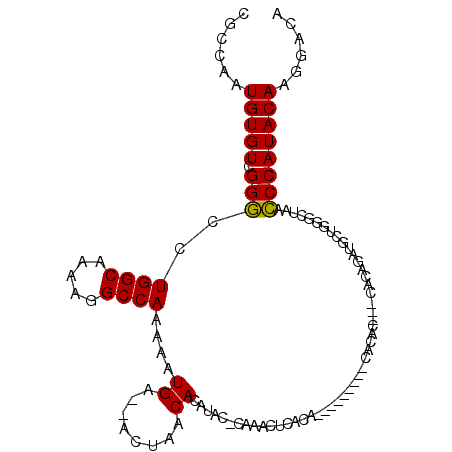

| Location | 11,596,490 – 11,596,590 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -28.88 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11596490 100 + 22407834 -----------UGUGAGUCUGCGAAUGUGUUAGU--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUU -----------.(((((.(((((....)).))))--))))..(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))). ( -34.50) >DroPse_CAF1 48000 79 + 1 CG----------------------------------CCAUUUUUGGCCUUUUGCCAGCCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGCU .(----------------------------------(((....))))........(((((.(((((((((.(.((..(((((......)))))..)).))))))))))))))) ( -30.90) >DroSec_CAF1 50639 108 + 1 UG---UGUGUGUGUUAGCCUUUGAAUGUGUUAGU--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUU ..---...(((..((((((.......).))))).--.)))..(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))). ( -35.00) >DroYak_CAF1 52618 104 + 1 UGAGC-------ACAUCCUUGUGGAUGUGUUAGU--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUU ..(((-------((((((....)))))))))...--......(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))). ( -44.90) >DroAna_CAF1 64580 108 + 1 UG---UGUGUGUGUGAGUGGC--UGUGUGUUAGUGCUCAUUUUUGGCCUUUUGCCAGGGCCGACACAUUGGCGUCCUUUUAUACGAAAAUGAGUUGAGGCAAUGUGUUGGGUU ..---.......(((((((.(--((.....))))))))))(((((((.....)))))))(((((((((((.(.((..(((((......)))))..)).))))))))))))... ( -34.40) >DroPer_CAF1 50480 79 + 1 CG----------------------------------CCAUUUUUGGCCUUUUGCCAGCCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGCU .(----------------------------------(((....))))........(((((.(((((((((.(.((..(((((......)))))..)).))))))))))))))) ( -30.90) >consensus UG__________GU_AG__U____AUGUGUUAGU__UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUU ..........................................(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))). (-28.88 = -29.38 + 0.50)



| Location | 11,596,490 – 11,596,590 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11596490 100 - 22407834 AACCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA--ACUAACACAUUCGCAGACUCACA----------- .....................(((((((..........(((((....))))).(((((......))))).)))))))--.......................----------- ( -19.20) >DroPse_CAF1 48000 79 - 1 AGCCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGGCUGGCAAAAGGCCAAAAAUGG----------------------------------CG (((((.(((((((((.((....................)).).))))))))..)))))........((((....)))----------------------------------). ( -22.85) >DroSec_CAF1 50639 108 - 1 AACCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA--ACUAACACAUUCAAAGGCUAACACACACA---CA ...((.(((((((((.((....................)).).)))))))).))(((((((.....))).....(((--(........)))).))))...........---.. ( -22.15) >DroYak_CAF1 52618 104 - 1 AACCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA--ACUAACACAUCCACAAGGAUGU-------GCUCA .....................(((((((..........(((((....))))).(((((......))))).)))))))--.....(((((((....))))))-------).... ( -29.20) >DroAna_CAF1 64580 108 - 1 AACCCAACACAUUGCCUCAACUCAUUUUCGUAUAAAAGGACGCCAAUGUGUCGGCCCUGGCAAAAGGCCAAAAAUGAGCACUAACACACA--GCCACUCACACACACA---CA ....................((((((((..........(((((....)))))((((.(.....).)))).))))))))............--................---.. ( -18.70) >DroPer_CAF1 50480 79 - 1 AGCCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGGCUGGCAAAAGGCCAAAAAUGG----------------------------------CG (((((.(((((((((.((....................)).).))))))))..)))))........((((....)))----------------------------------). ( -22.85) >consensus AACCCAACACAUUGCCUCAACUCAUUUUCGCAUAAAAGGACGCCAAUGUGUCGGGCCUGGCAAAAGGCCAAAAAUGA__ACUAACACAU____A__CU_AC__________CA ..(((.(((((((((.((....................)).).)))))))).)))..((((.....))))........................................... (-18.40 = -18.57 + 0.17)

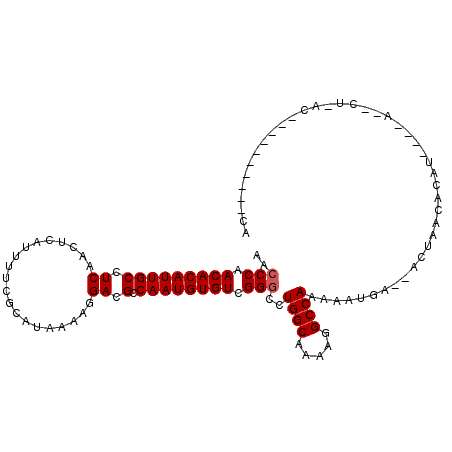
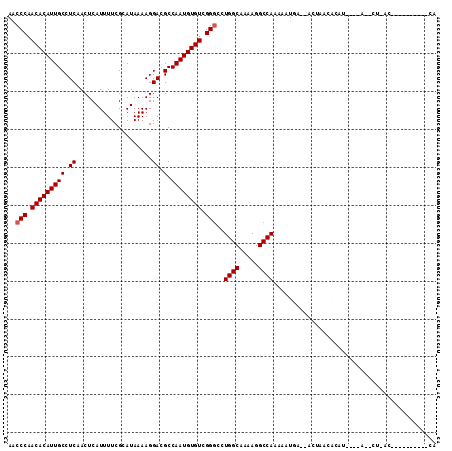
| Location | 11,596,512 – 11,596,615 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11596512 103 + 22407834 U--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUUUGCUAUUU-UUCGCAUGUCUU--------------UUUUC .--......(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))).(((.....-...)))......--------------..... ( -33.20) >DroPse_CAF1 48002 111 + 1 ---CCAUUUUUGGCCUUUUGCCAGCCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGCUAGCCAUUUUUUCCCAUCUCUCGAAGCUCU------CCCUC ---(((....)))......((.(((((.(((((((((.(.((..(((((......)))))..)).))))))))))))))).))..........................------..... ( -30.10) >DroEre_CAF1 81676 103 + 1 U--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUUUGCUAUUU-UUCGCAUGUCUU--------------UUUUC .--......(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))).(((.....-...)))......--------------..... ( -33.20) >DroYak_CAF1 52644 103 + 1 U--UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUUUGCUAUUU-UUCGCAUGUCUU--------------UUUUC .--......(((((.....)))))(((((((((((((.(.((..(((((......)))))..)).)))))))))))))).(((.....-...)))......--------------..... ( -33.20) >DroAna_CAF1 64608 119 + 1 UGCUCAUUUUUGGCCUUUUGCCAGGGCCGACACAUUGGCGUCCUUUUAUACGAAAAUGAGUUGAGGCAAUGUGUUGGGUUUGCUAUUU-UUCGUAUGUCUGCCCGCCCUUGCGCCAUUUC ..........((((.....((.(((((.(((((((((.(.((..(((((......)))))..)).))))))))))((((..(((((..-...))).))..))))))))).)))))).... ( -38.80) >DroPer_CAF1 50482 111 + 1 ---CCAUUUUUGGCCUUUUGCCAGCCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGCUAGCCAUUUUUUCCCAUCUCUCGAAGCUCU------CUUUU ---(((....)))......((.(((((.(((((((((.(.((..(((((......)))))..)).))))))))))))))).))..........................------..... ( -30.10) >consensus U__UCAUUUUUGGCCUUUUGCCAGGCCCGACACAUUGGCGUCCUUUUAUGCGAAAAUGAGUUGAGGCAAUGUGUUGGGUUUGCUAUUU_UUCGCAUGUCUU______________UUUUC ..........((((.....))))((((((((((((((.(.((..(((((......)))))..)).)))))))))))))))........................................ (-28.98 = -29.48 + 0.50)

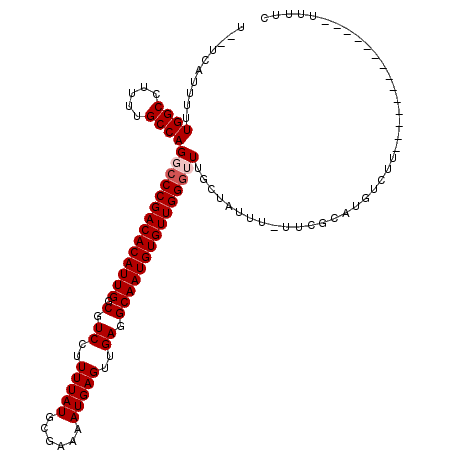
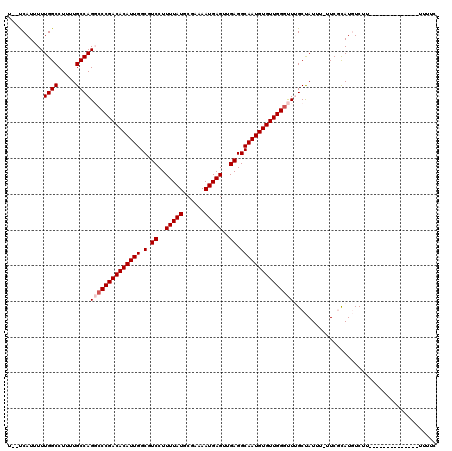
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:19 2006