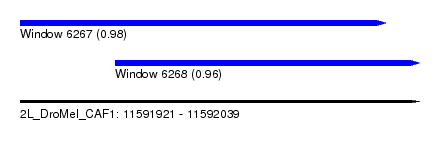
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,591,921 – 11,592,039 |
| Length | 118 |
| Max. P | 0.984437 |

| Location | 11,591,921 – 11,592,029 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11591921 108 + 22407834 AAU------------AUGUACUCAACCAGUUUUCCUUGCUGUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCGAGUUAUCUCAUUAGAUGUACUUGAGACUGUUGAG ...------------.....((((((((((.......))))......................................(((((.(((((((((....))))).))))))))).)))))) ( -20.90) >DroSec_CAF1 46026 120 + 1 AAUUCAAUACUCAACAAAUACUCAACCAGUUUUCCUCGCAGUCCAUUAUUAUACACCAUACACUCAAUUUUCGCCAGUUGUCUCCGAGUUAUCUCAUUAGAUGUACUUGAGACUGGCGAG ......................................................................(((((((((.....((((((((((....))))).))))).))))))))). ( -21.60) >DroSim_CAF1 48702 120 + 1 AAUUCAAUAUUCAUCAAACACUCAACCAGUUUUCCUCGCUGUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCAAAUUAUCUCAUUAGAUGUACUUGAGACUGGCGAG ..........................((((.......))))............................(((((((...(((((.....(((((....))))).....)))))))))))) ( -21.50) >DroEre_CAF1 77226 93 + 1 ------------------------ACCCAGUUCUUUUGCUGUCCAUUAUU--ACCCCAUACACUCAAUUCGCGCCAGUUGUCUCC-AGUUAUCUCAUUAGAUGUACUUGCGGCUGGCGAG ------------------------...((((......)))).........--...................((((((((((....-((((((((....))))).))).)))))))))).. ( -22.60) >DroYak_CAF1 47940 106 + 1 CA------------CAAGUACUCAACCAGCUAGUCUUGCAGCUCAUUAUU--ACCCCAUACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAGAUGUACUUGAGGCUGGCGAG ..------------((((.(((.........)))))))............--.................(((((((((...(((.(((((((((....))))).)))))))))))))))) ( -25.70) >DroAna_CAF1 60442 115 + 1 A--UCAAGG--UAAUGAAUAUUCAGGCUCUGUUCCUCACUCCUCGUCAUUAUCC-UUGCACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAUAUGUACUUGAGGUUGGCGAG .--.(((((--((((((......(((...((.....))..)))..)))))).))-)))...........((((((((...((((.(((((((........))).)))))))))))))))) ( -26.60) >consensus AA____________CAAAUACUCAACCAGUUUUCCUCGCUGUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAGAUGUACUUGAGACUGGCGAG .....................................................................((((((((((.....((((((((((....))))).))))).)))))))))) (-17.86 = -17.87 + 0.00)


| Location | 11,591,949 – 11,592,039 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11591949 90 + 22407834 GUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCGAGUUAUCUCAUUAGAUGUACUUGAGACUGUUGAGGUGGAAAAUU ...............((((...((((((...........(((((.(((((((((....))))).))))))))).))))))))))...... ( -19.40) >DroSec_CAF1 46066 90 + 1 GUCCAUUAUUAUACACCAUACACUCAAUUUUCGCCAGUUGUCUCCGAGUUAUCUCAUUAGAUGUACUUGAGACUGGCGAGGUGGAAAAUG .((((((..(((.....)))..........(((((((((.....((((((((((....))))).))))).)))))))))))))))..... ( -25.00) >DroSim_CAF1 48742 90 + 1 GUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCAAAUUAUCUCAUUAGAUGUACUUGAGACUGGCGAGGUGGAAAAUG .(((((...(((.....))).........(((((((...(((((.....(((((....))))).....)))))))))))))))))..... ( -23.40) >DroEre_CAF1 77242 87 + 1 GUCCAUUAUU--ACCCCAUACACUCAAUUCGCGCCAGUUGUCUCC-AGUUAUCUCAUUAGAUGUACUUGCGGCUGGCGAGGUGGAAAAUG .((((((...--...................((((((((((....-((((((((....))))).))).)))))))))).))))))..... ( -22.50) >DroYak_CAF1 47968 88 + 1 GCUCAUUAUU--ACCCCAUACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAGAUGUACUUGAGGCUGGCGAGGUGGAAAAUG ..........--...........((...((((((((((...(((.(((((((((....))))).)))))))))))))))))..))..... ( -26.20) >DroAna_CAF1 60478 89 + 1 CCUCGUCAUUAUCC-UUGCACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAUAUGUACUUGAGGUUGGCGAGGUGGAAAAUG ......((((.(((-(((......))).(((((((((...((((.(((((((........))).))))))))))))))))).))).)))) ( -20.50) >consensus GUCCAUUAUUAUACACCAUACACUCAAUUCUCGCCAGUUGUCUCCAAGUUAUCUCAUUAGAUGUACUUGAGACUGGCGAGGUGGAAAAUG .......................((...(((((((((((.....((((((((((....))))).))))).)))))))))))..))..... (-20.76 = -20.77 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:12 2006