| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,578,303 – 11,578,400 |
| Length | 97 |
| Max. P | 0.969484 |

| Location | 11,578,303 – 11,578,400 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -14.31 |
| Energy contribution | -16.37 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
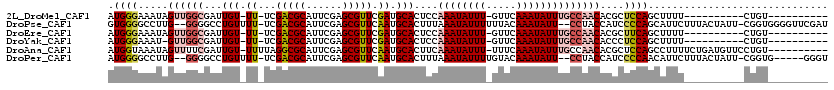
>2L_DroMel_CAF1 11578303 97 + 22407834 ----------ACAG----------AAAAGCUGGAGCGUGUUGGCAAAUAUUUGAAC-AAAUAUUUGGAGUGCAUCGAACGCUCGAAUGCGUCGA-AA-ACAAUCGCCAACUAUUUCCCAU ----------....----------.......((((...((((((((((((((....-))))))))...((...((((.(((......)))))))-..-))....))))))...))))... ( -27.10) >DroPse_CAF1 34656 114 + 1 AUCGAACCCCACCG-AAUAGUAAAGAAUGCUGGGGAUGGUAGG--AAUAUUUGUAAAAAAUAUUUAAAGUGCAUUGAACGCUCGAAUGCGUCGA-AAAACAGGCCCC--CAAGGCCCCAC ......(((((.((-............)).))))).(((...(--(((((((.....))))))))...((...((((.(((......)))))))-...)).((((..--...))))))). ( -26.60) >DroEre_CAF1 63307 97 + 1 ----------ACAG----------AAAAGCUGAAGCGUGUUGGCAAAUAUUUGAAC-AAAUAUUUGGAGUGCAUCGAACGCUCGAAUGCGUCGA-AA-ACAAUCGCCAACUAUUUCCCAU ----------...(----------(((.((....))..((((((((((((((....-))))))))...((...((((.(((......)))))))-..-))....))))))..)))).... ( -25.80) >DroYak_CAF1 33945 96 + 1 ----------ACAG----------AAAAGCUGGAGGGUGUUGGCAAAUAUUUGAAC-AAAUAUUUGGAGUGCAUCGAACGCUCGAAUGCGUCGA-AA-ACAAUCGCCAAC-AUUUCCCAU ----------....----------......(((..(((((((((((((((((....-))))))))...((...((((.(((......)))))))-..-))....))))))-)))..))). ( -32.60) >DroAna_CAF1 44617 108 + 1 ----------ACAGGAACAUCAGAAAAGGCUGGAGCGUGUUGGCAAAUAUUUGAAA-AAAUAUUUGAAGUGCAUUGAACGCUCGAAUGCGCCUAAAA-ACAAUCGAAAACUAUUUACCAU ----------..(((..((((((......)))((((((..((.(((((((((....-))))))))).....))....))))))..)))..)))....-...................... ( -23.30) >DroPer_CAF1 34548 109 + 1 ACCC-----CACCG-AAUAGUAAAGAAUGUUGGGGAUGGUAGG--AAUAUUUGUACAAAAUAUUUAAAGUGCAUUGAACGCUCGAAUGCGUCGA-AAAACAGGCCCC--CAAGGCCCCAU .(((-----((.((-............)).)))))((((...(--(((((((.....))))))))...((...((((.(((......)))))))-...)).((((..--...)))))))) ( -27.30) >consensus __________ACAG__________AAAAGCUGGAGCGUGUUGGCAAAUAUUUGAAC_AAAUAUUUGAAGUGCAUCGAACGCUCGAAUGCGUCGA_AA_ACAAUCGCCAACUAUUUCCCAU ...............................((((...((((((((((((((.....))))))))...((...((((.(((......)))))))....))....))))))...))))... (-14.31 = -16.37 + 2.06)


| Location | 11,578,303 – 11,578,400 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -14.13 |
| Energy contribution | -15.27 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11578303 97 - 22407834 AUGGGAAAUAGUUGGCGAUUGU-UU-UCGACGCAUUCGAGCGUUCGAUGCACUCCAAAUAUUU-GUUCAAAUAUUUGCCAACACGCUCCAGCUUUU----------CUGU---------- ((((((((..((((((((.(((-..-(((((((......))).)))).))).)).((((((((-....))))))))))))))..((....))))))----------))))---------- ( -29.20) >DroPse_CAF1 34656 114 - 1 GUGGGGCCUUG--GGGGCCUGUUUU-UCGACGCAUUCGAGCGUUCAAUGCACUUUAAAUAUUUUUUACAAAUAUU--CCUACCAUCCCCAGCAUUCUUUACUAUU-CGGUGGGGUUCGAU ((((((((...--..))))(((..(-(.(((((......))))).)).))).....(((((((.....)))))))--....))))(((((.(.............-.).)))))...... ( -26.14) >DroEre_CAF1 63307 97 - 1 AUGGGAAAUAGUUGGCGAUUGU-UU-UCGACGCAUUCGAGCGUUCGAUGCACUCCAAAUAUUU-GUUCAAAUAUUUGCCAACACGCUUCAGCUUUU----------CUGU---------- ((((((((..((((((((.(((-..-(((((((......))).)))).))).)).((((((((-....))))))))))))))..((....))))))----------))))---------- ( -29.20) >DroYak_CAF1 33945 96 - 1 AUGGGAAAU-GUUGGCGAUUGU-UU-UCGACGCAUUCGAGCGUUCGAUGCACUCCAAAUAUUU-GUUCAAAUAUUUGCCAACACCCUCCAGCUUUU----------CUGU---------- ..(((...(-((((((((.(((-..-(((((((......))).)))).))).)).((((((((-....))))))))))))))))))..........----------....---------- ( -29.60) >DroAna_CAF1 44617 108 - 1 AUGGUAAAUAGUUUUCGAUUGU-UUUUAGGCGCAUUCGAGCGUUCAAUGCACUUCAAAUAUUU-UUUCAAAUAUUUGCCAACACGCUCCAGCCUUUUCUGAUGUUCCUGU---------- .....((((((((...))))))-)).((((.((((..((((((.....(((.....(((((((-....))))))))))....))))))(((......))))))).)))).---------- ( -19.70) >DroPer_CAF1 34548 109 - 1 AUGGGGCCUUG--GGGGCCUGUUUU-UCGACGCAUUCGAGCGUUCAAUGCACUUUAAAUAUUUUGUACAAAUAUU--CCUACCAUCCCCAACAUUCUUUACUAUU-CGGUG-----GGGU .(((((...((--((((..(((..(-(.(((((......))))).)).))).....(((((((.....)))))))--))).))).))))).....(((((((...-.))))-----))). ( -25.30) >consensus AUGGGAAAUAGUUGGCGAUUGU_UU_UCGACGCAUUCGAGCGUUCAAUGCACUCCAAAUAUUU_GUUCAAAUAUUUGCCAACACGCUCCAGCUUUU__________CUGU__________ .((((.....((((((...(((.((...(((((......))))).)).)))....((((((((.....))))))))))))))....)))).............................. (-14.13 = -15.27 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:57 2006