| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,572,208 – 11,572,359 |
| Length | 151 |
| Max. P | 0.770136 |

| Location | 11,572,208 – 11,572,319 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11572208 111 + 22407834 CGCUUAAUUUCC-CAGCA-AACACUAAGUCAUUUCCGGGCAAACGGCCUG-GCCAUUACCUUGAUUAUCAUCUCAAGUUCGAGCCGGGCAUGGAAAUUCUUUUUC------CCACUCCGC .(((........-.))).-................((((......(((((-((......(((((........))))).....)))))))..(((((.....))))------)...)))). ( -26.10) >DroPse_CAF1 29128 105 + 1 CGCUUAAUUUCCCCAACAAAACACUAAGUCAUUUCCGGCCAAACGGCCUG-GCCAUUACCUAGAUUAUCAUUUCAAGUUCG------GCAUGGAAAUUU----UCCCUUUUGUCCU---- ...............((((((.........((((((((((.((((((...-)))........((........))..))).)------)).)))))))..----....))))))...---- ( -18.16) >DroSec_CAF1 26685 108 + 1 CGCUUAAUUUCC-CAGCA-AACACUAAGUCAUUUCCGGCCAAACGGCCG--GC--UAACCUUGAUUAUCAUCUCAAGUUCGAGCCGGUCAUGGAAAUUCUUUUUC------CCACUCCGC .(((........-.))).-................(((......(((((--((--(...(((((........)))))....))))))))..(((((.....))))------)....))). ( -27.70) >DroEre_CAF1 57413 106 + 1 CGCUUAAUUUCC-CAGCA-AACACUAAGUCAUUUCCGGCCAAACGGCCUA-GCCAUUACCUUGAUUAUCAUCUCAAGUUCGAGCCGGGCAUGGAAAUU-UUUUUC------CCACU---- .....(((((((-..((.-.((.....)).....((((((....(((...-))).....(((((........)))))...).)))))))..)))))))-......------.....---- ( -22.70) >DroAna_CAF1 38685 114 + 1 CGCUUAAUUUCC-CAGCA-AACACUAAGUCAUUUCCGGCCAAACGGCCUGGGCUAUUACCUUGAUUAUCAUCUCAAGUUUGAGUCAAGAAUGGAAAUUCAUUUUUUUUUUUGUACU---- ............-..(((-((....((((.((((((((((....))))..((((.....(((((........)))))....))))......))))))..)))).....)))))...---- ( -23.40) >DroPer_CAF1 28953 104 + 1 CGCUUAAUUUCCCCAACA-AACACUAAGUCAUUUCCGGCCAAACGGCCUG-GCCAUUACCUAGAUUAUCAUUUCAAGUUCG------GCAUGGAAAUUU----UCCCUUUUGUCCU---- ...............(((-((.........((((((((((.((((((...-)))........((........))..))).)------)).)))))))..----.....)))))...---- ( -17.69) >consensus CGCUUAAUUUCC_CAGCA_AACACUAAGUCAUUUCCGGCCAAACGGCCUG_GCCAUUACCUUGAUUAUCAUCUCAAGUUCGAGCCGGGCAUGGAAAUUCUUUUUC______CCACU____ .(((((..................))))).((((((((((....))))...........(((((........)))))..............))))))....................... (-13.90 = -14.40 + 0.50)

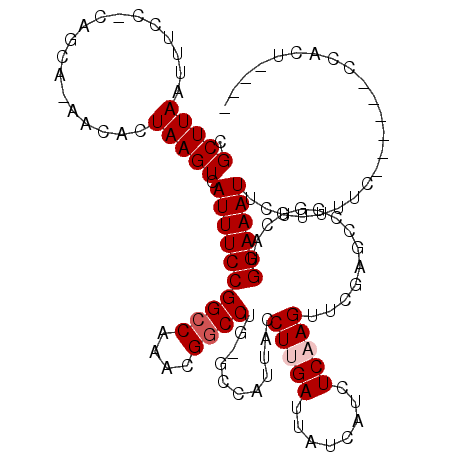

| Location | 11,572,246 – 11,572,359 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.23 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -24.32 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11572246 113 - 22407834 ACCUCAAACCGUGGUAAACAAACGGGAAAACGUGCCCCAUGCGGAGUGGGAAAAAGAAUUUCCAUGCCCGGCUCGAACUUGAGAUGAUAAUCAAGGUAAUGGCCAGGCCGUUU ........((((.........))))..(((((.(((......((.(((((((......)))))))..))((((....(((((........))))).....)))).)))))))) ( -34.10) >DroSec_CAF1 26723 110 - 1 ACCUCAAACCGAGGUAAACAAACGGGAAAACGUGCCCCAUGCGGAGUGGGAAAAAGAAUUUCCAUGACCGGCUCGAACUUGAGAUGAUAAUCAAGGUUA--GC-CGGCCGUUU (((((.....)))))........(((.........)))..((((.(((((((......)))))))..((((((....(((((........)))))...)--))-))))))).. ( -36.50) >DroSim_CAF1 26758 113 - 1 ACCUCAAACACAGGUAAACAAACGGGAAAACGUGCCCCAUGCGGAGUGGGAAAAAGAAUUUCCAUGACCGGCUCGAACUUGAGAUGAUAAUCAAGGUAAUGGCUAGGCCGUUU ((((.......))))............(((((.(((((((.(((.(((((((......)))))))..))).......(((((........)))))...))))...)))))))) ( -29.20) >DroEre_CAF1 57451 94 - 1 ACCUCAAGCCGAGGUAAACAAACGGG------------------AGUGGGAAAAA-AAUUUCCAUGCCCGGCUCGAACUUGAGAUGAUAAUCAAGGUAAUGGCUAGGCCGUUU ((((((((.(((((....)...((((------------------.(((((((...-..))))))).)))).))))..)))))(((....)))..)))((((((...)))))). ( -30.50) >DroYak_CAF1 27956 113 - 1 ACCUCAAGCCGAGGAAAACAAACGGGAGAACGUGCCCCCUGCGGAGUGGGAAAAAGAAUUUCCAUGCCCGGCUCGAACUUGAGAUGAUAAUCAAGGUAAUGGCUAGGCCGUUU .((((.....)))).............(((((.(((.....(((.(((((((......)))))))..)))(((....(((((........))))).....)))..)))))))) ( -32.90) >consensus ACCUCAAACCGAGGUAAACAAACGGGAAAACGUGCCCCAUGCGGAGUGGGAAAAAGAAUUUCCAUGCCCGGCUCGAACUUGAGAUGAUAAUCAAGGUAAUGGCUAGGCCGUUU (((((.....)))))....((((((....................(((((((......)))))))..((((((....(((((........))))).....)))).)))))))) (-24.32 = -25.20 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:53 2006