| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,569,071 – 11,569,186 |
| Length | 115 |
| Max. P | 0.872201 |

| Location | 11,569,071 – 11,569,186 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
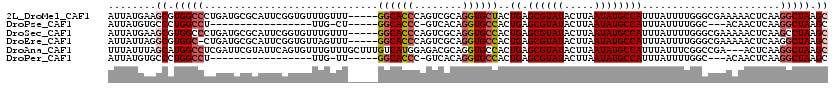
>2L_DroMel_CAF1 11569071 115 + 22407834 AUUAUGAAGCGUGGCCCUGAUGCGCAUUCGGUGUUUGUUU-----GGCACCCAGUCGCAGGUGCUACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGGCGAAAAACUCAAGGCUAAGC ........((.(((((.((((((((....((((((.....-----))))))((((.((....)).)))).))))).........((((..........))))......))).))))).)) ( -35.10) >DroPse_CAF1 25248 93 + 1 AUUAUGUGCCCUGGCCU-----------------UUG-CU-----GGCACCC-GUCACAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGC---ACAACUCAAGGCUAAGC .......((..((((((-----------------(((-((-----((((((.-......))))))..((.((((((.....))))))))........)))---)......))))))).)) ( -28.10) >DroSec_CAF1 23557 115 + 1 AUUAUGAAGCGUGGCCCUGAUGCGCAUUCGGUGUUUGUUU-----GGCACCCAGUCGCAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGGCGAAAAACUCAAGCCUAAGC ........((((((((((...(((.(((.((((((.....-----)))))).))))))))).)))))((.((((((.....))))))))......((((((...........)))))))) ( -37.40) >DroEre_CAF1 54175 114 + 1 AUUAUUAGGCGUGGC-CUGAUGCGCAUUCGGUGUUAGUUU-----GGCACCCAGUCGCAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGGCGAAAAACUCAAGGCUAAGC ........((.((((-((.((((((....((((((.....-----))))))((((.((....)).)))).))))))........((((..........)))).........)))))).)) ( -34.90) >DroAna_CAF1 35717 117 + 1 UUUAUUUAGCAUGGCCUCGAUUCGUAUUCAGUGUUUGUUUGCUUUGUCAUGGAGACGCAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUCGGCCGA---ACUCAAGGCUAAGC ........((.((((((.((((((...((((((...(..(((...(((.....))))))..)..))))))((((((.....)))))).............)))---).)).)))))).)) ( -31.80) >DroPer_CAF1 25416 93 + 1 AUUAUGUGCCCUGGCCU-----------------UUG-UU-----GGCACCC-GUCACAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGC---ACAACUCAAGGCUAAGC .......((..((((((-----------------(..-.(-----((((((.-......)))))))..(((.((((.....))))((((.......))))---....)))))))))).)) ( -28.10) >consensus AUUAUGAAGCGUGGCCCUGAUGCGCAUUCGGUGUUUGUUU_____GGCACCCAGUCGCAGGUGCCACUGAGCGUAUACUUAAUAUGCCAUUUAUUUUGGCCGAAAAACUCAAGGCUAAGC ........((.(((((.............................((((((........))))))..((.((((((.....)))))))).......................))))).)) (-19.14 = -19.87 + 0.72)

| Location | 11,569,071 – 11,569,186 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -16.61 |
| Energy contribution | -18.25 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11569071 115 - 22407834 GCUUAGCCUUGAGUUUUUCGCCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUAGCACCUGCGACUGGGUGCC-----AAACAAACACCGAAUGCGCAUCAGGGCCACGCUUCAUAAU ((...(((((((((..((((((..........)))............((((((((.((....)).))))))))..-----...........)))...)).)))))))...))........ ( -31.30) >DroPse_CAF1 25248 93 - 1 GCUUAGCCUUGAGUUGU---GCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGUGAC-GGGUGCC-----AG-CAA-----------------AGGCCAGGGCACAUAAU ((((.((((((((((((---((((.......)))))))..........))))..(((((((((...)-)))))))-----).-..)-----------------))))..))))....... ( -35.60) >DroSec_CAF1 23557 115 - 1 GCUUAGGCUUGAGUUUUUCGCCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGCGACUGGGUGCC-----AAACAAACACCGAAUGCGCAUCAGGGCCACGCUUCAUAAU ....((((.(((((.....(((..........))).......(....))))))(((((.((((((..(.((((..-----.......)))).)...)))...))))))))))))...... ( -30.20) >DroEre_CAF1 54175 114 - 1 GCUUAGCCUUGAGUUUUUCGCCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGCGACUGGGUGCC-----AAACUAACACCGAAUGCGCAUCAG-GCCACGCCUAAUAAU ((...((((.((((..((((((..........)))......(((...((((((((.((....)).))))))))..-----..)))......)))...)).))))-))...))........ ( -29.10) >DroAna_CAF1 35717 117 - 1 GCUUAGCCUUGAGU---UCGGCCGAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGCGUCUCCAUGACAAAGCAAACAAACACUGAAUACGAAUCGAGGCCAUGCUAAAUAAA ((...(((((((.(---(((((((.......))))...............((((((((....))(((.....)))............))))))...)))))))))))...))........ ( -33.90) >DroPer_CAF1 25416 93 - 1 GCUUAGCCUUGAGUUGU---GCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGUGAC-GGGUGCC-----AA-CAA-----------------AGGCCAGGGCACAUAAU ((((.((((((((((((---((((.......)))))))..........))))..(((((((((...)-)))))))-----).-..)-----------------))))..))))....... ( -35.60) >consensus GCUUAGCCUUGAGUUUUUCGCCCAAAAUAAAUGGCAUAUUAAGUAUACGCUCAGUGGCACCUGCGACUGGGUGCC_____AAACAAACACCGAAUGCGCAUCAAGGCCACGCCACAUAAU .....(((((((.........(((.......))).............((((((((.((....)).))))))))...........................)))))))............. (-16.61 = -18.25 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:49 2006