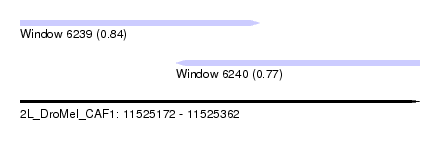
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,525,172 – 11,525,362 |
| Length | 190 |
| Max. P | 0.844362 |

| Location | 11,525,172 – 11,525,286 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -44.85 |
| Consensus MFE | -33.20 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
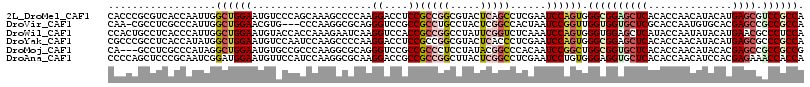
>2L_DroMel_CAF1 11525172 114 + 22407834 AGAUUCGUGUUGCUGUGCGUUUGGGUGGCAGCCGAAUAGCUAGGUGGAGCUGGCUUGUAUGCCGGGUUGUAGGCUGGCGGACGCUCAUGUAUGUUGGUGUGAGCUCCGCCCACU ..(((((.(((((..(.(....).)..)))))))))).....((((((.(((((......))))).)).......((((((.(((((..(......)..))))))))))))))) ( -49.20) >DroSim_CAF1 6524 114 + 1 AGGUUCGUGUUGCUGUGCGUUUGGGUGGCAGCCGAAUAACUGGGUGGAGCUGGCUUGUAAGCCGGGUUGUAGGCUGGCGGCCGCUCAUGUAUGUUGGUAUGAGCUCCGCCCACU ..(((((.(((((..(.(....).)..))))))))))...((((((((...((((.((.((((........)))).))))))(((((((........))))))))))))))).. ( -52.30) >DroEre_CAF1 6310 114 + 1 AGAUUCGUGUUGCCGUGCGUUUGGGUGGCGGCUGAAUAGCUGGGUGGAGCUGGCUUGUACGCCGGGUUGUAGGCUGGUGGUCGCUCAUGUAUGUUGGUGUGAGCUCCGCCCACU ..(((((.((((((((.(....).)))))))))))))((.(((((((((((.((.....((((((.(....).))))))...(((.((....)).))))).))))))))))))) ( -46.40) >DroWil_CAF1 38478 103 + 1 --AUUACUGUU------GGUAU---UGGCUGCCGAGUAACUCGGUGGCGGCGGUUUAUAUGAUGGAUUGUAUGCUGGAGGGCGUUCAUGUAUAUUGGUAUGAGCUCCACCCACU --.........------(((((---..((..(((((...)))))..)).(((((((((...))))))))))))))((..((.(((((((........))))))).))..))... ( -34.20) >DroYak_CAF1 6817 114 + 1 AGAUUCGUGUUGCUGUGCGUUUGUGUGGCGGCCGAGUAGCUGGGUGGAGCUGGCUUGUAUGCCGGAUUGUAGGCUGGCGGGCGCUCAUGUAUGUUGGUGUGAGCUCCGCCCACU ..(((((.(((((((..(....)..))))))))))))((.(((((((((((.(((.....(((........))).))).)))(((((..(......)..))))))))))))))) ( -52.50) >DroAna_CAF1 2772 111 + 1 AGAUUCGUGUU---GUGCGUCUGGGUGGCAGCAGGAUAACUGGGUGGUGCCGGUCUGUACGAUGGAUUGUAGGCUGGUGGUUUCUCGUGGAUGUUGGUGUGAGCACCUCCCACA .......((((---.((((((.....))).))).))))...(((.(.(((((((((..((((....))))))))))))).)..)))((((.....((((....))))..)))). ( -34.50) >consensus AGAUUCGUGUUGCUGUGCGUUUGGGUGGCAGCCGAAUAACUGGGUGGAGCUGGCUUGUAUGCCGGAUUGUAGGCUGGCGGGCGCUCAUGUAUGUUGGUGUGAGCUCCGCCCACU ..(((((.(((((((..(....)..))))))))))))...(((((((((((((((((((........)))))))))))....(((((((........))))))))))))))).. (-33.20 = -34.02 + 0.81)

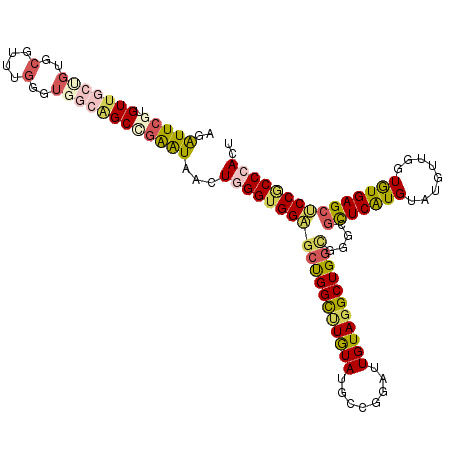
| Location | 11,525,246 – 11,525,362 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.99 |
| Mean single sequence MFE | -40.11 |
| Consensus MFE | -22.07 |
| Energy contribution | -23.21 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11525246 116 - 22407834 CACCCGCGUCACCAAUUGGCUGGAAUGUCCCAGCAAAGCCCCAAGGACCUCCGCCGGCGUACUCAGCCUCGAAUCCAGUGGGCGGAGCUCACACCAACAUACAUGAGCGUCCGCCA (((.....((.......(((.(((..((((..............)))).))))))(((.......)))..)).....)))(((((((((((............))))).)))))). ( -37.74) >DroVir_CAF1 29676 112 - 1 CAA-CGCCUCGCCCAUUGGCUGGAACGUG---CCCAAGGCGCAGGGUCCGCCGCCUGCCUACUCGGCCACUAAUCCGGUUGGUGGUGCUCGCACCAAUGUGCACGAGCCGCCGCCA ...-((.(((((((((..((((((..(((---.((.((..(((((........)))))...)).)).)))...))))))..)))).))..((((....))))..))).))...... ( -46.20) >DroWil_CAF1 38541 116 - 1 CCACUGCCUCACCCAUUGGCUGGAAUGUACCACCAAAGAAUCAAGGUCCACCGCCGGCCUAUUCGGUCUCAAAUCCAGUGGGUGGAGCUCAUACCAAUAUACAUGAACGCCCUCCA .....((..((((((((((((((..((.(((.............))).))...)))))).....((........))))))))))..))............................ ( -32.22) >DroYak_CAF1 6891 116 - 1 CGCCCGCCUCACCAUAUGGCUGGAAUGUCCAAUCCAAGCCCCAAGGACCUCCGCCGGCGUACUCACCCUCGAAUCCAGUGGGCGGAGCUCACACCAACAUACAUGAGCGCCCGCCA ((((.((..........(((((((........))).))))....((....)))).))))....(((...........)))(((((.(((((............)))))..))))). ( -36.20) >DroMoj_CAF1 31360 113 - 1 CA---GCCUCGCCCAUAGGCUGGAAUGUGCCGCCCAAGGCGCAGGGUCCGCCGCCCUCCUAUACGGCCCACAAUCCGGCUGGCGGUGCUCACACCAACAUACACGAGCCGCCGCCG .(---(((..(((....))).(((.((((((......))))))(((.(((.............))))))....)))))))((((((((((..............))).))))))). ( -48.16) >DroAna_CAF1 2843 116 - 1 CCCCAGCUCCCGCAAUCGGAUGGAAUGUUCCAUCCAAGGCGCAAGGACCGCCGCCGGCUUACUCGGCCUCGAAUCCUGUGGGAGGUGCUCACACCAACAUCCACGAGAAACCACCA ...((.((((((((...(((((((....)))))))..((((.......))))(((((.....))))).........)))))))).))(((..............)))......... ( -40.14) >consensus CACCCGCCUCACCAAUUGGCUGGAAUGUCCCACCCAAGGCGCAAGGACCGCCGCCGGCCUACUCGGCCUCGAAUCCAGUGGGCGGAGCUCACACCAACAUACACGAGCGCCCGCCA ..................((((((....................((....))(((((.....)))))......)))))).((((((((((..............)))).)))))). (-22.07 = -23.21 + 1.14)
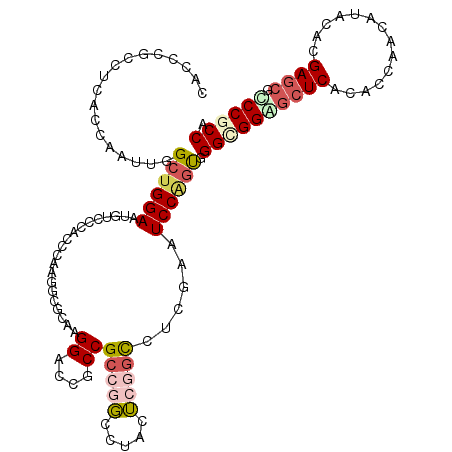
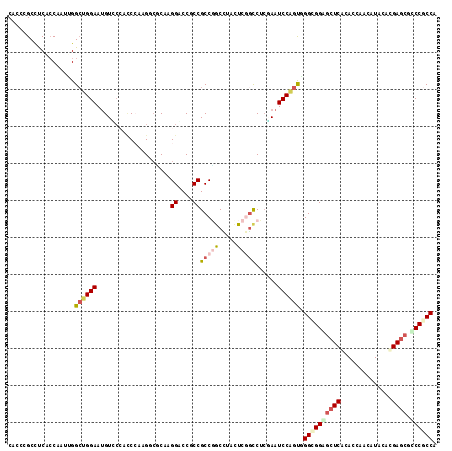
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:44 2006