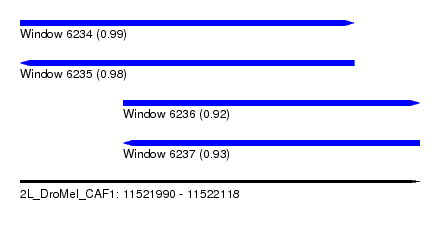
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,521,990 – 11,522,118 |
| Length | 128 |
| Max. P | 0.988575 |

| Location | 11,521,990 – 11,522,097 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -48.72 |
| Consensus MFE | -32.24 |
| Energy contribution | -33.24 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11521990 107 + 22407834 -UAUAGAGAGCUGGAAAUUUGUUUUCUGGGAGGC------------UUGUAUGCCGGCGGUCUUUCGCGAAUGUUCGUGGGUGCUCCACCCACGGUAUCAUGAGCUGAGUAAGCCGGCGG -..((((((((.........))))))))......------------.....(((((((...(((..((..(((.(((((((((...)))))))))...)))..)).)))...))))))). ( -43.20) >DroSec_CAF1 3928 119 + 1 -UACAGAGAUCCGGAAUUUGGUUUUCUGUGUGGCUGGUCGCGGGUAUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGG -((((((((.(((.....))).))))))))((((.((((((.(((((...))))).)))))).((((((.(((.(((((((((...)))))))))...))).))).)))...)))).... ( -53.90) >DroSim_CAF1 3581 119 + 1 -UACAGAGAUCCGGAAUUCGGUUUUCUGUGUGGCUUGUCGCGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGG -((((((((.(((.....))).))))))))..((..(((....)))..)).((((((((..(((..(((.(((.(((((((((...)))))))))...))).))).)))..)))))))). ( -56.70) >DroEre_CAF1 2959 119 + 1 -UAUAGAGAGCUGGAACUUUGCUUACUGGAUGGCUUGUUCAGUGGUGUGUAGGCCGGCGGUCUCUCGCGAAUGUUCGUGGGAGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGG -((((.(..((((.(((...(((........)))..)))))))..).)))).(((((((..(((..((..(((.(((((((.......)))))))...)))..)).)))..))))))).. ( -48.60) >DroYak_CAF1 3639 105 + 1 AUAUAGGGAGCU---------------GGAUGCCUUUUUUUGGGGCUCGUAGGCCGGCGGAGUUUCGCGAAUGUUCGUGCGAGCUCCACUUACAGUAUCAUGAGCUGAGUACGCCGGCGG ..........((---------------(((.(((((.....))))))).)))((((((((((((((((((....))))).))))))).....((((.......)))).....)))))).. ( -41.20) >consensus _UAUAGAGAGCUGGAAUUUGGUUUUCUGGGUGGCUUGUCGCGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUCGUGGGUGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGG ..........((((((.......)))))).......................(((((((....(((((..(((.(((((((((...)))))))))...)))..)).)))..))))))).. (-32.24 = -33.24 + 1.00)



| Location | 11,521,990 – 11,522,097 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -24.88 |
| Energy contribution | -26.80 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11521990 107 - 22407834 CCGCCGGCUUACUCAGCUCAUGAUACCGUGGGUGGAGCACCCACGAACAUUCGCGAAAGACCGCCGGCAUACAA------------GCCUCCCAGAAAACAAAUUUCCAGCUCUCUAUA- ..((((((...((..((..(((....((((((((...))))))))..)))..))...))...))))))......------------.................................- ( -29.40) >DroSec_CAF1 3928 119 - 1 CCGCCGGCGUACUCAGCGCAUGAUACCGUGGGUGGAGCACCCACAAACAUUCGCGAGAGGCCGCCGGCAUACAAUACCCGCGACCAGCCACACAGAAAACCAAAUUCCGGAUCUCUGUA- ..(((((((..(((.(((.(((.....(((((((...)))))))...))).)))..)))..)))))))...........((.....))...(((((...((.......))...))))).- ( -45.20) >DroSim_CAF1 3581 119 - 1 CCGCCGGCGUACUCAGCGCAUGAUACCGUGGGUGGAGCACCCACAAACAUUCGCGAGAGGCCGCCGGCAUACAAGCCCCGCGACAAGCCACACAGAAAACCGAAUUCCGGAUCUCUGUA- ..(((((((..(((.(((.(((.....(((((((...)))))))...))).)))..)))..)))))))...........((.....))...(((((...(((.....)))...))))).- ( -47.90) >DroEre_CAF1 2959 119 - 1 CCGCCGGCGUACUCAGCUCAUGAUACCGUGGGUGGAGCUCCCACGAACAUUCGCGAGAGACCGCCGGCCUACACACCACUGAACAAGCCAUCCAGUAAGCAAAGUUCCAGCUCUCUAUA- ..(((((((..(((.((..(((....((((((.......))))))..)))..))..)))..))))))).........................((..(((.........)))..))...- ( -34.90) >DroYak_CAF1 3639 105 - 1 CCGCCGGCGUACUCAGCUCAUGAUACUGUAAGUGGAGCUCGCACGAACAUUCGCGAAACUCCGCCGGCCUACGAGCCCCAAAAAAAGGCAUCC---------------AGCUCCCUAUAU ..(((((((((.(((.....)))))).......((((.((((..........))))..))))))))))....((((((........)).....---------------.))))....... ( -29.50) >consensus CCGCCGGCGUACUCAGCUCAUGAUACCGUGGGUGGAGCACCCACGAACAUUCGCGAGAGACCGCCGGCAUACAAGCCCCGCAACAAGCCACCCAGAAAACAAAAUUCCAGCUCUCUAUA_ ..(((((((..(((.((..(((....((((((((...))))))))..)))..))..)))..))))))).................................................... (-24.88 = -26.80 + 1.92)


| Location | 11,522,023 – 11,522,118 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -35.12 |
| Energy contribution | -35.44 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11522023 95 + 22407834 ------UUGUAUGCCGGCGGUCUUUCGCGAAUGUUCGUGGGUGCUCCACCCACGGUAUCAUGAGCUGAGUAAGCCGGCGGAGGUCCUUUGGGCAAAU---------------UGUG ------.....(((((((...(((..((..(((.(((((((((...)))))))))...)))..)).)))...)))))))...((((...))))....---------------.... ( -39.80) >DroSec_CAF1 3967 101 + 1 CGGGUAUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGGAGGACCUUUGGGCAAAU---------------UGUG .((((......((((((((..(((..(((.(((.(((((((((...)))))))))...))).))).)))..))))))))....))))..........---------------.... ( -45.30) >DroSim_CAF1 3620 101 + 1 CGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUUGUGGGUGCUCCACCCACGGUAUCAUGCGCUGAGUACGCCGGCGGAGGACCUUUGGGCAAAU---------------UGUG .(((((((...((((((((..(((..(((.(((.(((((((((...)))))))))...))).))).)))..)))))))).))).)))).........---------------.... ( -45.60) >DroEre_CAF1 2998 116 + 1 AGUGGUGUGUAGGCCGGCGGUCUCUCGCGAAUGUUCGUGGGAGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGGAGGUCCUUUGGGCAAGUUAGAUCCACCUCCAGUGCG ........(((.(((((((..(((..((..(((.(((((((.......)))))))...)))..)).)))..)))))))((((((.....((((......).)))))))))..))). ( -48.50) >DroYak_CAF1 3664 101 + 1 UGGGGCUCGUAGGCCGGCGGAGUUUCGCGAAUGUUCGUGCGAGCUCCACUUACAGUAUCAUGAGCUGAGUACGCCGGCGGAGGUCCUUUGGGCGAA--------------AG-UCU .(((.(((....((((((((((((((((((....))))).))))))).....((((.......)))).....)))))).))).)))...((((...--------------.)-))) ( -45.00) >consensus CGGGGCUUGUAUGCCGGCGGCCUCUCGCGAAUGUUCGUGGGUGCUCCACCCACGGUAUCAUGAGCUGAGUACGCCGGCGGAGGUCCUUUGGGCAAAU_______________UGUG ......((((..(((((((....(((((..(((.(((((((((...)))))))))...)))..)).)))..)))))))((....)).....))))..................... (-35.12 = -35.44 + 0.32)



| Location | 11,522,023 – 11,522,118 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11522023 95 - 22407834 CACA---------------AUUUGCCCAAAGGACCUCCGCCGGCUUACUCAGCUCAUGAUACCGUGGGUGGAGCACCCACGAACAUUCGCGAAAGACCGCCGGCAUACAA------ ....---------------...........((....))((((((...((..((..(((....((((((((...))))))))..)))..))...))...))))))......------ ( -31.10) >DroSec_CAF1 3967 101 - 1 CACA---------------AUUUGCCCAAAGGUCCUCCGCCGGCGUACUCAGCGCAUGAUACCGUGGGUGGAGCACCCACAAACAUUCGCGAGAGGCCGCCGGCAUACAAUACCCG ....---------------...........((....))(((((((..(((.(((.(((.....(((((((...)))))))...))).)))..)))..)))))))............ ( -39.70) >DroSim_CAF1 3620 101 - 1 CACA---------------AUUUGCCCAAAGGUCCUCCGCCGGCGUACUCAGCGCAUGAUACCGUGGGUGGAGCACCCACAAACAUUCGCGAGAGGCCGCCGGCAUACAAGCCCCG ....---------------....((.....((....))(((((((..(((.(((.(((.....(((((((...)))))))...))).)))..)))..)))))))......)).... ( -40.30) >DroEre_CAF1 2998 116 - 1 CGCACUGGAGGUGGAUCUAACUUGCCCAAAGGACCUCCGCCGGCGUACUCAGCUCAUGAUACCGUGGGUGGAGCUCCCACGAACAUUCGCGAGAGACCGCCGGCCUACACACCACU ......((.(((((.(((..(((((.....(((.(((((((.(((...(((.....)))...))).))))))).)))...((....)))))))))))))))..))........... ( -41.80) >DroYak_CAF1 3664 101 - 1 AGA-CU--------------UUCGCCCAAAGGACCUCCGCCGGCGUACUCAGCUCAUGAUACUGUAAGUGGAGCUCGCACGAACAUUCGCGAAACUCCGCCGGCCUACGAGCCCCA ...-..--------------..........((..(((.(((((((((.(((.....)))))).......((((.((((..........))))..))))))))))....)))..)). ( -30.60) >consensus CACA_______________AUUUGCCCAAAGGACCUCCGCCGGCGUACUCAGCUCAUGAUACCGUGGGUGGAGCACCCACGAACAUUCGCGAGAGACCGCCGGCAUACAAGCCCCG .......................(((....(((.(((((((.(((...(((.....)))...))).))))))).)))...........(((......))).)))............ (-27.32 = -27.68 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:41 2006