
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,487,517 – 11,487,859 |
| Length | 342 |
| Max. P | 0.999988 |

| Location | 11,487,517 – 11,487,614 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.64 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -35.98 |
| Energy contribution | -35.66 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487517 97 + 22407834 CCUCGAAGGAGAACGAAAAAGUGGCCCGGAA------UAGCGGGGCGGCU----CAAAGUGGGUGGUGGCCAAAGGGUGAAC-----CACCCAAGCGGCAACAUUGCUGACG .(((....)))........(((.((((.(..------...).)))).)))----.....((((((((.(((....)))..))-----)))))).((((((....))))).). ( -40.50) >DroSec_CAF1 51358 97 + 1 CCUCGAAGGAGAGCGAAAAAGUGGCCCGGAA------AAGCGGGGCGGCU----CAAAGUGGGUGGUGGCCAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACG .(((....)))(((((...(((.((((.(..------...).)))).)))----.....((((((((.(((....))).).)-----))))))...(....).))))).... ( -39.90) >DroSim_CAF1 56186 97 + 1 UCUCGAAGGAGAGCGAAAAAGUGGCCCGGAA------AAGCGGGGCGGCU----CAUAGUGGGUGGUGGCCAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACG ((((....))))((.....(((.((((.(..------...).)))).)))----.....((((((((.(((....))).).)-----)))))).))((((....)))).... ( -40.90) >DroEre_CAF1 57038 107 + 1 CCUCGAAGGAGAGCGAAAAAGUGGCCCGGAAAAGGAAAAGCGGGGCGGCACCCAGUGGGUGGGUGGUGGUUAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACG .(((....)))(((((....((.((((.(...........).)))).)).((...(((((((((.(...........).)))-----))))))...)).....))))).... ( -39.30) >DroYak_CAF1 57095 102 + 1 CCUCCGAGGAGAGCGAAAAAGUGGCCCGGAA------AAGCGGGGCGGCU----GAAAGUGGGUGGUGGCCAAAGGGUGACCCGAGCCACCCAAGCGGCAACAUUGCUGACG .(((....)))(((((.....((.((((...------...)))).))(((----(....((((((((.......((....))...))))))))..))))....))))).... ( -41.90) >consensus CCUCGAAGGAGAGCGAAAAAGUGGCCCGGAA______AAGCGGGGCGGCU____CAAAGUGGGUGGUGGCCAAAGGGUGACC_____CACCCAAGCGGCAACAUUGCUGACG .(((....))).........((.((((.(...........).)))).)).........((.((..((((((...(((((........)))))....)))..)))..)).)). (-35.98 = -35.66 + -0.32)



| Location | 11,487,550 – 11,487,644 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487550 94 + 22407834 GCGGGGCGGCU----CAAAGUGGGUGGUGGCCAAAGGGUGAAC-----CACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG (((...((((.----.....((((((((.(((....)))..))-----))))))...(....)...)))).)))............(((((.......))))) ( -31.80) >DroSec_CAF1 51391 94 + 1 GCGGGGCGGCU----CAAAGUGGGUGGUGGCCAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG (((...((((.----.....((((((((.(((....))).).)-----))))))...(....)...)))).)))............(((((.......))))) ( -30.70) >DroSim_CAF1 56219 94 + 1 GCGGGGCGGCU----CAUAGUGGGUGGUGGCCAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG (((...((((.----.....((((((((.(((....))).).)-----))))))...(....)...)))).)))............(((((.......))))) ( -30.70) >DroEre_CAF1 57077 98 + 1 GCGGGGCGGCACCCAGUGGGUGGGUGGUGGUUAAAGGGUGACC-----CACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG ..(((......)))....((((..(((((((....(((((...-----)))))..((((((....))))).))))))))..)))).(((((.......))))) ( -36.40) >DroYak_CAF1 57128 99 + 1 GCGGGGCGGCU----GAAAGUGGGUGGUGGCCAAAGGGUGACCCGAGCCACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG ....((.(..(----((.((((.(.((((((....((....))...)))))))...(((((....))))).)))).)))..).)).(((((.......))))) ( -34.50) >consensus GCGGGGCGGCU____CAAAGUGGGUGGUGGCCAAAGGGUGACC_____CACCCAAGCGGCAACAUUGCUGACGCUAUCAAACACCACAAACACAUUUAGUUUG ((......)).........(((..(((((((....(((((........)))))..((((((....))))).))))))))..)))..(((((.......))))) (-30.46 = -30.30 + -0.16)


| Location | 11,487,614 – 11,487,705 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.20 |
| Mean single sequence MFE | -20.01 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487614 91 - 22407834 GCAAAUUGCCAGAAGAAAAA---GAAGUAGCAAGAAAA------GUGAGCGACGCAAAAACAAAUUC-CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG--------- ((...((((...........---......)))).....------((.....)))).....(((((..-((((((((.......)))))))).)))))....--------- ( -16.43) >DroSec_CAF1 51455 91 - 1 GCAAAUUGUCAGAAGAAAAA---GAAGCAGCAAGAAAA------GUGAGCGACGCAAAAACAAAUUC-CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG--------- ......(((((((.......---...((.((.......------....))...))............-((((((((.......))))))))..))))))).--------- ( -17.50) >DroSim_CAF1 56283 91 - 1 GCAAAUUGCCAGAAGAAAAA---GAAGCAGCAAGAAAA------GUGAGCGACGCAAAAACAAAUUC-CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG--------- ((....(((...........---...)))((.......------....))...)).....(((((..-((((((((.......)))))))).)))))....--------- ( -16.64) >DroEre_CAF1 57145 94 - 1 GCAAAUUGCCAAAAGAAGAAGCAGAGGCAGCAAGAAAA------GUGAGCGACGCAAAAACAAAUUC-CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG--------- ((....((((...............))))((.......------....))...)).....(((((..-((((((((.......)))))))).)))))....--------- ( -20.36) >DroYak_CAF1 57197 88 - 1 GCAAAUUGCCAAAAGAAGAAGC------AGCAAGAAAA------GUGAGCGACGCAAAAACAAAUUC-CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG--------- ((....(((...........))------)((.......------....))...)).....(((((..-((((((((.......)))))))).)))))....--------- ( -16.90) >DroPer_CAF1 55627 97 - 1 G-AAAGUGCGAGUAGA---------G---GCACAAAAACCUUUGGGGAGCGAGGCAAAAACAAAUUCGCCGCAAACUAAAUGUGUUUGUGGCAUUUGAUAUGCCCCCGGG .-...((((.......---------.---)))).....(((..((((.(((((...........)))(((((((((.......))))))))).........))))))))) ( -32.20) >consensus GCAAAUUGCCAGAAGAAAAA___GAAGCAGCAAGAAAA______GUGAGCGACGCAAAAACAAAUUC_CCGCAAACUAAAUGUGUUUGUGGUGUUUGAUAG_________ .....((((....................))))...........................(((((...((((((((.......)))))))).)))))............. (-14.87 = -14.32 + -0.55)


| Location | 11,487,670 – 11,487,781 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487670 111 + 22407834 --UUUUCUUGCUACUUC---UUUUUCUUCUGGCAAUUUGCGCCAUAAAAAUGUGAAAAA-AAAAUUUGCAA--AAAAUAAGAGCGGAAUAAAAAU-GCCAUGAGAGGCAAGAAGAAACUG --((((((((((.....---....((((.(((((..((.((((.((....((..((...-....))..)).--....)).).))).))......)-)))).))))))))))))))..... ( -24.70) >DroSec_CAF1 51511 113 + 1 --UUUUCUUGCUGCUUC---UUUUUCUUCUGACAAUUUGCGCCAUAAAAAUGUGAAAAAAAAAAUUUGCAAC-AAAAUAAGAGCGGAAUAAAAAU-GCCAUGAGAAGCAAGAAGAAAUUG --.((((((..((((((---((.............(((((..........((..((........))..))..-.........)))))........-.....))))))))..))))))... ( -20.37) >DroSim_CAF1 56339 112 + 1 --UUUUCUUGCUGCUUC---UUUUUCUUCUGGCAAUUUGCGCCAUAAAAAUGUGAAAAA-AAAAUUUGCAAC-AAAAUAAGAGCGGAAUAAAAAU-GCCAUGAGAGGCAAGAAGAAAUUG --((((((((((.....---....((((.(((((..((.(((((((....)))).....-...((((.....-.))))....))).))......)-)))).))))))))))))))..... ( -24.40) >DroEre_CAF1 57201 112 + 1 --UUUUCUUGCUGCCUCUGCUUCUUCUUUUGGCAAUUUGCGCCAUAAAAAUGUGCAAA----AAAUUGCAAC-AAAAUAAAAGCGGUAUAAAAAA-GCCAUGAGAGGCAAGAAGAACCUG --..(((((..(((((((((((.((.((((((((((((..(((((....))).))...----)))))))..)-)))).))))))(((........-)))...)))))))..))))).... ( -33.40) >DroYak_CAF1 57253 107 + 1 --UUUUCUUGCU------GCUUCUUCUUUUGGCAAUUUGCGCCAUAAAAAUGUGCAAAA---AAAUUGCAAC-AAAAUAAAAGCGGAAUAAAAAG-GCCAUGAGAGGCAAGAAGAAACUG --.((((((..(------(((((((....((((...((.(((........(((((((..---...)))).))-)........))).)).......-)))).))))))))..))))))... ( -28.49) >DroPer_CAF1 55697 104 + 1 GGUUUUUGUGC---C---------UCUACUCGCACUUU-CGCCAUCAAAAUGUGGAAAA-GAAA--UGAAAUAAGAAUAAGAGAUACUCCAAAAGAUCGAUUAUGCUGAUGAAAAGUCUA (..((((((((---.---------.......))))(((-(.((((......))))....-))))--.......((.((((..(((.((.....)))))..)))).))....))))..).. ( -14.30) >consensus __UUUUCUUGCUGCUUC___UUUUUCUUCUGGCAAUUUGCGCCAUAAAAAUGUGAAAAA_AAAAUUUGCAAC_AAAAUAAGAGCGGAAUAAAAAU_GCCAUGAGAGGCAAGAAGAAACUG ..((((((((((............((((.((((.((((....((((....))))..........(((((.............)))))....)))).)))).))))))))))))))..... (-15.36 = -16.17 + 0.81)


| Location | 11,487,670 – 11,487,781 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487670 111 - 22407834 CAGUUUCUUCUUGCCUCUCAUGGC-AUUUUUAUUCCGCUCUUAUUUU--UUGCAAAUUUU-UUUUUCACAUUUUUAUGGCGCAAAUUGCCAGAAGAAAAA---GAAGUAGCAAGAAAA-- ...........((((......)))-).................((((--((((..(((((-((((((.........(((((.....)))))...))))))---))))).)))))))).-- ( -23.50) >DroSec_CAF1 51511 113 - 1 CAAUUUCUUCUUGCUUCUCAUGGC-AUUUUUAUUCCGCUCUUAUUUU-GUUGCAAAUUUUUUUUUUCACAUUUUUAUGGCGCAAAUUGUCAGAAGAAAAA---GAAGCAGCAAGAAAA-- ...((((((.(((((((.....((-(.....................-..)))...((((((((((.(((.(((........))).))).))))))))))---))))))).)))))).-- ( -21.90) >DroSim_CAF1 56339 112 - 1 CAAUUUCUUCUUGCCUCUCAUGGC-AUUUUUAUUCCGCUCUUAUUUU-GUUGCAAAUUUU-UUUUUCACAUUUUUAUGGCGCAAAUUGCCAGAAGAAAAA---GAAGCAGCAAGAAAA-- ...........((((......)))-).................((((-(((((...((((-((((((.........(((((.....))))))))))))))---)..)))))))))...-- ( -25.70) >DroEre_CAF1 57201 112 - 1 CAGGUUCUUCUUGCCUCUCAUGGC-UUUUUUAUACCGCUUUUAUUUU-GUUGCAAUUU----UUUGCACAUUUUUAUGGCGCAAAUUGCCAAAAGAAGAAGCAGAGGCAGCAAGAAAA-- ....(((((.((((((((...((.-.........))((((((.((((-(..(((((((----..(((.(((....)))))).))))))))))))..)))))))))))))).)))))..-- ( -34.10) >DroYak_CAF1 57253 107 - 1 CAGUUUCUUCUUGCCUCUCAUGGC-CUUUUUAUUCCGCUUUUAUUUU-GUUGCAAUUU---UUUUGCACAUUUUUAUGGCGCAAAUUGCCAAAAGAAGAAGC------AGCAAGAAAA-- ..(((((((((((((......)))-..........((((.......(-((.((((...---..))))))).......))))...........))))))))))------..........-- ( -26.94) >DroPer_CAF1 55697 104 - 1 UAGACUUUUCAUCAGCAUAAUCGAUCUUUUGGAGUAUCUCUUAUUCUUAUUUCA--UUUC-UUUUCCACAUUUUGAUGGCG-AAAGUGCGAGUAGA---------G---GCACAAAAACC ..............((......((......((((((.....))))))....)).--.(((-(.((((((.(((((....))-)))))).))).)))---------)---))......... ( -15.80) >consensus CAGUUUCUUCUUGCCUCUCAUGGC_AUUUUUAUUCCGCUCUUAUUUU_GUUGCAAAUUUU_UUUUUCACAUUUUUAUGGCGCAAAUUGCCAGAAGAAAAA___GAAGCAGCAAGAAAA__ .......(((((((......................((.............)).................(((((.(((((.....))))).)))))............))))))).... (-10.70 = -11.62 + 0.92)


| Location | 11,487,742 – 11,487,859 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487742 117 + 22407834 GAGCGGAAUAAAAAUGCCAUGAGAGGCAAGAAGAAACUGUCCGACAAAUAAAAAUGGCAUAAAAAUAUUCCACUCACCUGAGAGGUGCGAAUUGGGUCCCCAAUACACUGUCAUAUA (((.((((((...(((((((..(.((((.........)))))...........))))))).....)))))).)))...(((..((((...(((((....))))).)))).))).... ( -29.52) >DroSec_CAF1 51585 117 + 1 GAGCGGAAUAAAAAUGCCAUGAGAAGCAAGAAGAAAUUGUCCGACAAAUAAAAAUGGCAUAAAAAUAUUCCACUCACCUGAGAGGUGCGAAUUGGGUACCCAAUACACGGUCAUAGA (((.((((((...(((((((.....((((.......)))).............))))))).....)))))).)))..(((.((.(((...(((((....))))).)))..)).))). ( -26.67) >DroSim_CAF1 56412 117 + 1 GAGCGGAAUAAAAAUGCCAUGAGAGGCAAGAAGAAAUUGUCCGACAAAUAAAAAUGGCAUAAAAAUAUUCCACUCACCUGAGAGGUGCGAAUUGGGUCCCCAAUACACUGUCAUAGA (((.((((((...(((((((..(.(((((.......))))))...........))))))).....)))))).)))..(((.((((((...(((((....))))).)))).)).))). ( -31.72) >DroEre_CAF1 57274 117 + 1 AAGCGGUAUAAAAAAGCCAUGAGAGGCAAGAAGAACCUGUCCGACAAAUAAAAAUGGCAUAAAAAUAUUCCACUUACCUCAGAGGUGCGAAUCGGGUGCCCAAUACACUGCGAGAUA ..(((((........(((......)))............................(((((......(((((((((.......))))).))))...)))))......)))))...... ( -24.00) >DroYak_CAF1 57321 117 + 1 AAGCGGAAUAAAAAGGCCAUGAGAGGCAAGAAGAAACUGUCCGACGAAUAAAAAUGGCAUAAAAAUAUUCCACUCACCUCAGAGGUGCGAAUCGGGUCUCCAAUACACUGUCGAGUA ....((((((.....(((((..(.((((.........)))))...........))))).......))))))((((....(((.(((....)))((....))......)))..)))). ( -20.62) >consensus GAGCGGAAUAAAAAUGCCAUGAGAGGCAAGAAGAAACUGUCCGACAAAUAAAAAUGGCAUAAAAAUAUUCCACUCACCUGAGAGGUGCGAAUUGGGUCCCCAAUACACUGUCAUAUA (((.((((((...(((((((((.((...........)).))............))))))).....)))))).)))(((.....)))..(.(((((....))))).)........... (-20.22 = -20.78 + 0.56)

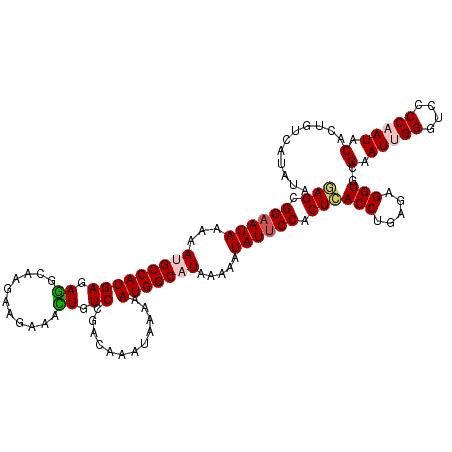

| Location | 11,487,742 – 11,487,859 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -30.84 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.50 |
| SVM RNA-class probability | 0.999988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11487742 117 - 22407834 UAUAUGACAGUGUAUUGGGGACCCAAUUCGCACCUCUCAGGUGAGUGGAAUAUUUUUAUGCCAUUUUUAUUUGUCGGACAGUUUCUUCUUGCCUCUCAUGGCAUUUUUAUUCCGCUC .........(((.(((((....))))).)))(((.....)))((((((((((.....(((((((........((.(((........))).)).....)))))))...)))))))))) ( -33.62) >DroSec_CAF1 51585 117 - 1 UCUAUGACCGUGUAUUGGGUACCCAAUUCGCACCUCUCAGGUGAGUGGAAUAUUUUUAUGCCAUUUUUAUUUGUCGGACAAUUUCUUCUUGCUUCUCAUGGCAUUUUUAUUCCGCUC .........(((.(((((....))))).)))(((.....)))((((((((((.....(((((((........((.(((........))).)).....)))))))...)))))))))) ( -34.42) >DroSim_CAF1 56412 117 - 1 UCUAUGACAGUGUAUUGGGGACCCAAUUCGCACCUCUCAGGUGAGUGGAAUAUUUUUAUGCCAUUUUUAUUUGUCGGACAAUUUCUUCUUGCCUCUCAUGGCAUUUUUAUUCCGCUC .........(((.(((((....))))).)))(((.....)))((((((((((.....(((((((........((.(((........))).)).....)))))))...)))))))))) ( -33.62) >DroEre_CAF1 57274 117 - 1 UAUCUCGCAGUGUAUUGGGCACCCGAUUCGCACCUCUGAGGUAAGUGGAAUAUUUUUAUGCCAUUUUUAUUUGUCGGACAGGUUCUUCUUGCCUCUCAUGGCUUUUUUAUACCGCUU (((((((..(((((((((....)))))..))))...)))))))(((((.(((.......(((((............((.((((.......)))).))))))).....))).))))). ( -30.10) >DroYak_CAF1 57321 117 - 1 UACUCGACAGUGUAUUGGAGACCCGAUUCGCACCUCUGAGGUGAGUGGAAUAUUUUUAUGCCAUUUUUAUUCGUCGGACAGUUUCUUCUUGCCUCUCAUGGCCUUUUUAUUCCGCUU ..((((...(((((((((....)))))..))))...))))..((((((((((.......(((((........((.(((........))).)).....))))).....)))))))))) ( -28.12) >consensus UAUAUGACAGUGUAUUGGGGACCCAAUUCGCACCUCUCAGGUGAGUGGAAUAUUUUUAUGCCAUUUUUAUUUGUCGGACAGUUUCUUCUUGCCUCUCAUGGCAUUUUUAUUCCGCUC .........(((.(((((....))))).)))(((.....)))((((((((((.....(((((((........((.(((........))).)).....)))))))...)))))))))) (-30.84 = -30.80 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:25 2006