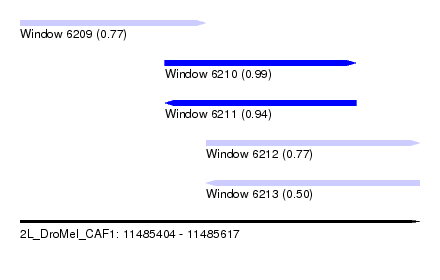
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,485,404 – 11,485,617 |
| Length | 213 |
| Max. P | 0.994667 |

| Location | 11,485,404 – 11,485,503 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11485404 99 + 22407834 GUCCACCUCCACCA---CCGCCCCAUUAUCCACCACCAGGGGGAUAUCUGUAUGGUGGCCACCAUUGGCAGGGAUAUCCUCCUCCGCCACCUCCGCCU------------------CCUU ...........(((---(((..(...(((((.((....)).)))))...)..)))))).......((((.((((.....))))..)))).........------------------.... ( -26.50) >DroSec_CAF1 49226 99 + 1 GUCCACCUCCACCA---CCGCCCCAUUAUCCACCACCGGGGGGAUAUCCGUAUGGUGGCCACUAUUGGCAGGGAUAUCCUCCUCCGCCACCUCCGCCU------------------CCUU ...........(((---(((..(...(((((.((....)).)))))...)..)))))).......((((.((((.....))))..)))).........------------------.... ( -26.40) >DroSim_CAF1 53008 99 + 1 GUCCACCUCCACCA---CCGCCCCACUAUCCACCACCAGGGGGAUAUCCGUAUGGUGGCCACUAUUGGCAGGGAUAUCCCCCUCCGCCACCACCUCCU------------------CCUU ...........(((---(((..(...(((((.((....)).)))))...)..)))))).......((((((((......))))..)))).........------------------.... ( -27.30) >DroYak_CAF1 54880 120 + 1 GUCCACCUCCACCACCACCACCCCAUUGUCUACCACCGGGGGGAUAUCCGCAUGCUGGUCACUAUUGGCAUGGUCAUCCUCCUCCGCCAACUCCGCCAACUCCGCCGCCUCCGGUACCAU ..................................(((((((((....))((..(.((((.....(((((..((.......))...)))))....)))).)...))...)))))))..... ( -26.20) >consensus GUCCACCUCCACCA___CCGCCCCAUUAUCCACCACCAGGGGGAUAUCCGUAUGGUGGCCACUAUUGGCAGGGAUAUCCUCCUCCGCCACCUCCGCCU__________________CCUU .....................................((((((((((((........((((....))))..))))))))))))..................................... (-21.51 = -21.20 + -0.31)


| Location | 11,485,481 – 11,485,583 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -27.76 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11485481 102 + 22407834 CCUCCGCCACCUCCGCCU------------------CCUUCUUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGCGGUUGCCCAGGAGGCGGUCCACCUCAA ............((((((------------------(((..................((..((((....))))..)).(((((.((((...)))).)))))))))))))).......... ( -38.40) >DroSec_CAF1 49303 102 + 1 CCUCCGCCACCUCCGCCU------------------CCUUCAUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAA ............((((((------------------(((..................((..((((....))))..)).(((((.((((...)))).)))))))))))))).......... ( -36.50) >DroSim_CAF1 53085 102 + 1 CCUCCGCCACCACCUCCU------------------CCUUCAUCACACUACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGCCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAA ........(((.((((((------------------.....................((..((((....))))..)).((((..((((...))))..)))))))))).)))......... ( -33.50) >DroYak_CAF1 54960 120 + 1 CCUCCGCCAACUCCGCCAACUCCGCCGCCUCCGGUACCAUCACCACAUCACCCGCAACCAUCAGGAUCUCCACCACGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAG .......................((((((((((((......))).........((((((((..((((..((.......))..)))).....))))))))...)))))))))......... ( -41.40) >consensus CCUCCGCCACCUCCGCCU__________________CCUUCAUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAA ...(((((.((..............................................((..((((....))))..)).((((..((((...))))..)))).)).))))).......... (-27.76 = -28.58 + 0.81)


| Location | 11,485,481 – 11,485,583 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -46.45 |
| Consensus MFE | -35.99 |
| Energy contribution | -37.17 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11485481 102 - 22407834 UUGAGGUGGACCGCCUCCUGGGCAACCGCAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAAGAAGG------------------AGGCGGAGGUGGCGGAGG .....((.(.(((((((((...((.((((...)))).((((((((..((((....)))).....))))))))...))....)))------------------))))))...).))..... ( -48.10) >DroSec_CAF1 49303 102 - 1 UUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGG------------------AGGCGGAGGUGGCGGAGG .....((.(.(((((((((((....)).((.((((..((((((((..((((....)))).....)))))))).)))).)).)))------------------))))))...).))..... ( -48.70) >DroSim_CAF1 53085 102 - 1 UUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGGCACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUAGUGUGAUGAAGG------------------AGGAGGUGGUGGCGGAGG .....((.(.(((((((((((....)).((...((.(((((((((..((((....)))).....))))))).)).)).))....------------------)))))))))).))..... ( -45.80) >DroYak_CAF1 54960 120 - 1 CUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCGUGGUGGAGAUCCUGAUGGUUGCGGGUGAUGUGGUGAUGGUACCGGAGGCGGCGGAGUUGGCGGAGUUGGCGGAGG ........(.((((((((.((...((((((..(((..(((((((((.(.((....))).))))....))))).)))..)).)))).)))))))))))...((..((...))..))..... ( -43.20) >consensus UUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUGAAGG__________________AGGCGGAGGUGGCGGAGG ........(.(((((((((((....)))((.((((..((((((((..((((....)))).....)))))))).)))).))..........................)))))))))..... (-35.99 = -37.17 + 1.19)


| Location | 11,485,503 – 11,485,617 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11485503 114 + 22407834 CUUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGCGGUUGCCCAGGAGGCGGUCCACCUCAAAGUGGAUUUCCUGGUACCGAAAGAC------CUCACAACG ......(((.....((.((..((((....))))..)).(((((.((((...)))).))))).))(((..((((((......))))))..))))))..(....)..------......... ( -39.10) >DroSec_CAF1 49325 114 + 1 CAUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAAAGUGGAUCUCCCGGUACCGAAAGAC------CUCACAACG ......(((.....((.((..((((....))))..)).(((((.((((...)))).))))).)).((.(((((((......))))))).)).)))..(....)..------......... ( -37.40) >DroSim_CAF1 53107 114 + 1 CAUCACACUACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGCCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAAAAUGGAUCUCCCGGUACCGAAAGAC------CUCACAACG ........((((..((.((..((((....))))..)).((((..((((...))))..)))).)).((.((((((........)))))).)).)))).(....)..------......... ( -35.10) >DroYak_CAF1 55000 120 + 1 CACCACAUCACCCGCAACCAUCAGGAUCUCCACCACGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAGAGUGGAUCGCCUGGUACCGAAAGACACCCAACUCCCAACG .............((((((((..((((..((.......))..)))).....))))))))...(((((((((((((......)))))))))))(((..(....)..)))......)).... ( -45.50) >consensus CAUCACACCACCCCCCACCAUCUGGAUCUCCAGCAGGAGGGUGUCCGCAUUGUGGUUGCCCAGGAGGCGGUCCACCUCAAAGUGGAUCUCCCGGUACCGAAAGAC______CUCACAACG ......(((..((....((..((((....))))..)).((((..((((...))))..)))).)).((.(((((((......))))))).)).)))..(....)................. (-32.22 = -32.48 + 0.25)


| Location | 11,485,503 – 11,485,617 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -48.53 |
| Consensus MFE | -39.45 |
| Energy contribution | -40.07 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11485503 114 - 22407834 CGUUGUGAG------GUCUUUCGGUACCAGGAAAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCGCAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAAG (((((((((------....)))(((.((((((..((((((....)))))).....))))))...)))))))))....((((((((..((((....)))).....))))))))........ ( -45.80) >DroSec_CAF1 49325 114 - 1 CGUUGUGAG------GUCUUUCGGUACCGGGAGAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUG (((((((.(------((((..((....))..)))((((((....))))))))(((.....)))...)))))))....((((((((..((((....)))).....))))))))........ ( -51.00) >DroSim_CAF1 53107 114 - 1 CGUUGUGAG------GUCUUUCGGUACCGGGAGAUCCAUUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGGCACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUAGUGUGAUG (((((((.(------((((..((....))..)))((((((....))))))))(((.....)))...)))))))((.(((((((((..((((....)))).....))))))).)).))... ( -48.60) >DroYak_CAF1 55000 120 - 1 CGUUGGGAGUUGGGUGUCUUUCGGUACCAGGCGAUCCACUCUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCGUGGUGGAGAUCCUGAUGGUUGCGGGUGAUGUGGUG (((..(((((..(((..(....)..)))..))..((((((.(((((((..((((....(((....)))....)))).)).))))).))))))..)))..)))(..(((.....)))..). ( -48.70) >consensus CGUUGUGAG______GUCUUUCGGUACCAGGAGAUCCACUUUGAGGUGGACCGCCUCCUGGGCAACCACAAUGCGGACACCCUCCUGCUGGAGAUCCAGAUGGUGGGGGGUGGUGUGAUG (((((((((..........)))(((.(((((((.((((((....))))))....)))))))...)))))))))((.(((((((((..((((....)))).....))))))).)).))... (-39.45 = -40.07 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:18 2006