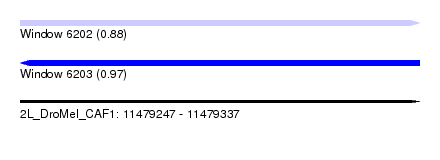
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,479,247 – 11,479,337 |
| Length | 90 |
| Max. P | 0.974859 |

| Location | 11,479,247 – 11,479,337 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
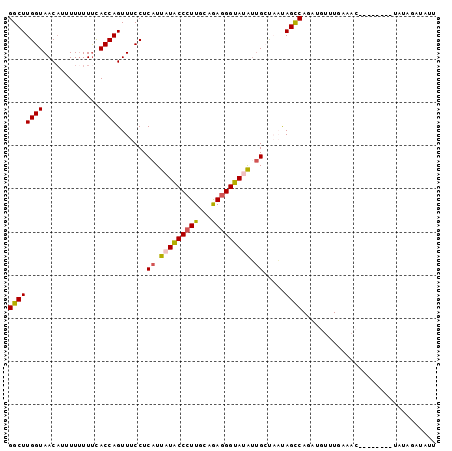
>2L_DroMel_CAF1 11479247 90 + 22407834 GGCUUGGUAACAUUUUUUUUUACCAGUUUCCUCAUUAUACCCUUGCAGAGGGUAUAUUGCUAAUAGCCAGAUGUUUGAAA---------UAAAGAUAUU ((((((((((.........)))))).......((.(((((((((...))))))))).)).....)))).(((((((....---------...))))))) ( -22.90) >DroSec_CAF1 43044 98 + 1 GGCUUGGUAACAUUUU-UUUCACCAGUUUCCUCAUUAUACCCUUACAAAGGGUAUAUUGCUAAUAGCCAGAUGUUUGAAACGCAGUGAAUAUAGAUAUU ...........((((.-.(((((..(((((..(((((((((((.....)))))))...((.....))..))))...)))))...)))))...))))... ( -21.50) >DroSim_CAF1 46774 90 + 1 GGCUUGGUAACAUUUUUUUUCACCAGUUUCCUCAUUAUACCAUUACAAAGGGUAUAUUGCUAAUAGCCAGUUGUUUGAAAC--------UAUAGCUAU- ((((((((.............)))).......((.((((((.........)))))).)).....))))((((((.......--------.))))))..- ( -16.22) >DroEre_CAF1 49075 78 + 1 GGCUUGGUAACAAUUUUUUCUACCAGUUUCCUC-UUCUACCCUUGCUGAGGGUAUGUCGAUUUCAGUCAGAAUAUUGCA-------------------- (((((((((...........)))))).......-...(((((((...))))))).)))(((....)))...........-------------------- ( -14.20) >DroYak_CAF1 48862 98 + 1 GGCUUGGUAACCAUUUUUUUCACCAGUUUCCUCAUUCUGCCCUUGCUGAGGGUAUAAUGACUUUAGUCAGCAGUUUGCAACGCAGUGAAU-UACAUAUU ((((((((.............))))......(((((.(((((((...))))))).)))))....)))).(.((((..(......)..)))-).)..... ( -20.52) >consensus GGCUUGGUAACAUUUUUUUUCACCAGUUUCCUCAUUAUACCCUUGCAGAGGGUAUAUUGCUAAUAGCCAGAUGUUUGAAAC________UAUAGAUAUU ((((((((.............)))).......((.(((((((((...))))))))).)).....))))............................... (-14.90 = -14.90 + 0.00)


| Location | 11,479,247 – 11,479,337 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -13.96 |
| Energy contribution | -15.16 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11479247 90 - 22407834 AAUAUCUUUA---------UUUCAAACAUCUGGCUAUUAGCAAUAUACCCUCUGCAAGGGUAUAAUGAGGAAACUGGUAAAAAAAAAUGUUACCAAGCC ..........---------............((((..(((((.((((((((.....))))))))((.((....)).)).........)))))...)))) ( -21.60) >DroSec_CAF1 43044 98 - 1 AAUAUCUAUAUUCACUGCGUUUCAAACAUCUGGCUAUUAGCAAUAUACCCUUUGUAAGGGUAUAAUGAGGAAACUGGUGAAA-AAAAUGUUACCAAGCC ..........(((((((.(((((...(((...((.....))..((((((((.....)))))))))))..)))))))))))).-................ ( -23.10) >DroSim_CAF1 46774 90 - 1 -AUAGCUAUA--------GUUUCAAACAACUGGCUAUUAGCAAUAUACCCUUUGUAAUGGUAUAAUGAGGAAACUGGUGAAAAAAAAUGUUACCAAGCC -...(((..(--------(((((...((....(((((((.(((........))))))))))....))..))))))(((((.........))))).))). ( -18.80) >DroEre_CAF1 49075 78 - 1 --------------------UGCAAUAUUCUGACUGAAAUCGACAUACCCUCAGCAAGGGUAGAA-GAGGAAACUGGUAGAAAAAAUUGUUACCAAGCC --------------------.(((((.(((((.((..........((((((.....))))))...-.((....)))))))))...)))))......... ( -15.20) >DroYak_CAF1 48862 98 - 1 AAUAUGUA-AUUCACUGCGUUGCAAACUGCUGACUAAAGUCAUUAUACCCUCAGCAAGGGCAGAAUGAGGAAACUGGUGAAAAAAAUGGUUACCAAGCC ........-.((((((..........((((((((....))))......(((.....)))))))....((....))))))))......((((....)))) ( -20.30) >consensus AAUAUCUAUA________GUUUCAAACAUCUGGCUAUUAGCAAUAUACCCUCUGCAAGGGUAUAAUGAGGAAACUGGUGAAAAAAAAUGUUACCAAGCC ...............................((((.....((.((((((((.....)))))))).))((....))(((((.........))))).)))) (-13.96 = -15.16 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:08 2006