
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,471,628 – 11,471,731 |
| Length | 103 |
| Max. P | 0.968192 |

| Location | 11,471,628 – 11,471,731 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -9.49 |
| Energy contribution | -10.64 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
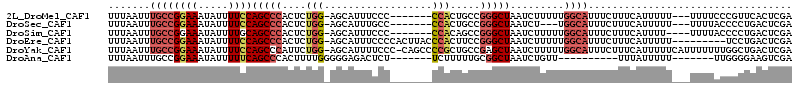
>2L_DroMel_CAF1 11471628 103 + 22407834 UUUAAUUUGCCGGAAAUAUUUUCCAGCCCACUCUGG-AGCAUUUCCC-------CCACUGCCGGGCUAAUCUUUUUGGCAUUUCUUUCAUUUUU---UUUUCCCGUUCACUCGA .......((((((((((...((((((......))))-)).))))))(-------((......)))...........))))..............---................. ( -20.80) >DroSec_CAF1 35172 100 + 1 UUUAAUUUGCCGGAAAUAUUUUCCAGCCCACUCUGG-AGCAUUUGCC-------CCACUGCCGGGCUAAUCU---UGGCAUUUCUUUCAUUUUU---UUUUACCCCUGACUCGA .......((((((((.....))))(((((.(..(((-.((....)).-------)))..)..))))).....---.))))..............---................. ( -24.20) >DroSim_CAF1 37783 102 + 1 UUUAAUUUGCCGGAAAUAUUUUGCAGCCCACUCUGG-AGCAUUUCCC-------CCACAGCCGGGCUAAUCUUUUUGGCAUUUCUUUCAUUUU----UUUUACCCCUGACUCGA .......(((((((((.(((....(((((.((.(((-..........-------))).))..)))))))).))))))))).............----................. ( -21.00) >DroEre_CAF1 35751 104 + 1 UUUAAUUUGCCGGAAAUAUUUUCCAGCCCACUCUGG-AGCAUUUCCCCACUUACCCACUUCCGGGCUAAUCUUUUUGGCAUUUCUUUCAUUUUU---------UCCUGACUCGA .......((((((((((...((((((......))))-)).)))))).......(((......)))...........))))..............---------........... ( -21.00) >DroYak_CAF1 36207 112 + 1 UUUAAUUUGCCGGAAAUAUUUUCCAGCCCAUUCUGG-AGCAUUUUCCC-CAGCCCCGCUGCCGAGCUAAUCUUUUUGGCAUUUCUUUCAUUUUUCAUUUUUUUGGCUGACUCGA ...........(((((....((((((......))))-))...))))).-(((((.((....)).(((((.....)))))........................)))))...... ( -23.00) >DroAna_CAF1 33099 90 + 1 UUUAAUUUGCCGGAAAUAUUUUUCAGCCCACUUUUGGGGGAGACUCU-------UCUUUUUGCGGCUAAUCUGUU----------UUUAUUUUU-------UUGGGGAAGUCGA .(..((((.((.((((....))))((((((.....((((....))))-------......)).))))........----------.........-------...)).))))..) ( -18.50) >consensus UUUAAUUUGCCGGAAAUAUUUUCCAGCCCACUCUGG_AGCAUUUCCC_______CCACUGCCGGGCUAAUCUUUUUGGCAUUUCUUUCAUUUUU___UUUUACCCCUGACUCGA .......((((((((.....))))(((((....(((..................))).....))))).........)))).................................. ( -9.49 = -10.64 + 1.14)
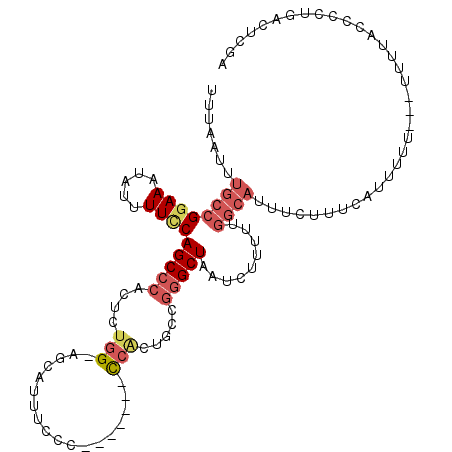
| Location | 11,471,628 – 11,471,731 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -10.39 |
| Energy contribution | -12.31 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
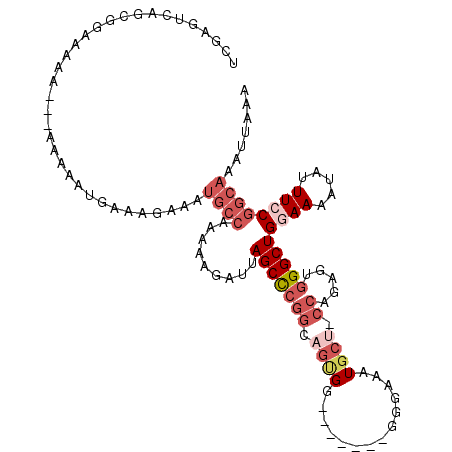
>2L_DroMel_CAF1 11471628 103 - 22407834 UCGAGUGAACGGGAAAA---AAAAAUGAAAGAAAUGCCAAAAAGAUUAGCCCGGCAGUGG-------GGGAAAUGCU-CCAGAGUGGGCUGGAAAAUAUUUCCGGCAAAUUAAA ((..(....)..))...---..............((((.........((((((.(..(((-------((......))-)))..)))))))((((.....))))))))....... ( -26.60) >DroSec_CAF1 35172 100 - 1 UCGAGUCAGGGGUAAAA---AAAAAUGAAAGAAAUGCCA---AGAUUAGCCCGGCAGUGG-------GGCAAAUGCU-CCAGAGUGGGCUGGAAAAUAUUUCCGGCAAAUUAAA ((...(((.........---.....)))..))..((((.---.....((((((.(..(((-------(((....)))-)))..)))))))((((.....))))))))....... ( -30.34) >DroSim_CAF1 37783 102 - 1 UCGAGUCAGGGGUAAAA----AAAAUGAAAGAAAUGCCAAAAAGAUUAGCCCGGCUGUGG-------GGGAAAUGCU-CCAGAGUGGGCUGCAAAAUAUUUCCGGCAAAUUAAA ....(((.(..(((...----....((..(....)..)).......(((((((.((.(((-------((......))-))).))))))))).....)))..).)))........ ( -28.00) >DroEre_CAF1 35751 104 - 1 UCGAGUCAGGA---------AAAAAUGAAAGAAAUGCCAAAAAGAUUAGCCCGGAAGUGGGUAAGUGGGGAAAUGCU-CCAGAGUGGGCUGGAAAAUAUUUCCGGCAAAUUAAA ((...(((...---------.....)))..))..((((.......(((.(((....).)).)))....(((((((.(-((((......)))))...)))))))))))....... ( -27.90) >DroYak_CAF1 36207 112 - 1 UCGAGUCAGCCAAAAAAAUGAAAAAUGAAAGAAAUGCCAAAAAGAUUAGCUCGGCAGCGGGGCUG-GGGAAAAUGCU-CCAGAAUGGGCUGGAAAAUAUUUCCGGCAAAUUAAA ....(((((((.......................((((..............)))).((...(((-(((......))-))))..))))))((((.....)))))))........ ( -25.84) >DroAna_CAF1 33099 90 - 1 UCGACUUCCCCAA-------AAAAAUAAA----------AACAGAUUAGCCGCAAAAAGA-------AGAGUCUCCCCCAAAAGUGGGCUGAAAAAUAUUUCCGGCAAAUUAAA .............-------.........----------....((((.((((.(((....-------....((...((((....))))..))......))).)))).))))... ( -12.54) >consensus UCGAGUCAGCGGAAAAA___AAAAAUGAAAGAAAUGCCAAAAAGAUUAGCCCGGCAGUGG_______GGGAAAUGCU_CCAGAGUGGGCUGGAAAAUAUUUCCGGCAAAUUAAA ..................................((((.........(((((((.((((..............)))).)).....)))))((((.....))))))))....... (-10.39 = -12.31 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:04 2006