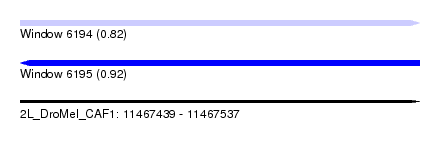
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,467,439 – 11,467,537 |
| Length | 98 |
| Max. P | 0.920506 |

| Location | 11,467,439 – 11,467,537 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -19.23 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11467439 98 + 22407834 ------------CUGGCCUUUACCAAUUAUUGCAAUGCGCUCUGCGCACGCACCUCCGGCAAUUAAAAUUAAUUUC-CUCGCAGACGCCAUUGUCUGGCGCAAAAGCCAGA ------------.(((......)))......((((((.((((((((....)......((.(((((....))))).)-)..))))).))))))))(((((......))))). ( -28.90) >DroPse_CAF1 31611 101 + 1 UGGCCUCUGCAUCUGGCCUCUGCCAAUUAUUGUAGUG-------CGCACGCACCUCCGGCAAUUAAAAUUAAUUUCCCUCGCAGACGCCAUUGUCUGGCGG---GGCCCAG .((((.((((...((((.(((((...........(((-------(....))))....((.(((((....))))).))...))))).)))).......))))---))))... ( -34.60) >DroEre_CAF1 31503 98 + 1 ------------CUGGCCUUUGCCAAUUAUUGCAACGCGCUCUGCGCACGCACCUCCGGCAAUUAAAAUUAAUUUC-CUCGCAGACGCCAUUGUCUGGCGCAAAAGCCAGU ------------(((((.(((((......((((...((((...))))..........((.(((((....))))).)-)..))))..((((.....))))))))).))))). ( -31.90) >DroYak_CAF1 31891 98 + 1 ------------CUGGCCUUUGCCAAUUAUUGCAAUGCGCUCUGCGCACGCACCUCCGGCAAUUAAAAUUAAUUUC-CUCGCAGACGCCAUUGUCUGGCGCAAAAGCCAGA ------------(((((.(((((......((((..(((((...))))).........((.(((((....))))).)-)..))))..((((.....))))))))).))))). ( -32.80) >DroAna_CAF1 29473 102 + 1 --------AGAUCUGGCUUUUGCCAAUUAUUGCAAUGCGCAUUGCGCACGCACCUCGGGCAAUUAAAAUUAAUUUC-CUAGCAGACGCCAUUGUCUGGCGCAAGAGACAGG --------....(((.(((((((......((((..(((((...)))))........(((.(((((....))))).)-)).))))..((((.....))))))))))).))). ( -31.20) >DroPer_CAF1 32616 95 + 1 UGGCCUC------UGGCCUCUGCCAAUUAUUGUAGUG-------CGCACGCACCUCCGGCAAUUAAAAUUAAUUUCCCUCGCAGACGCCAUUGUCUGGCGG---GGCCCGG .((((((------((((.(((((...........(((-------(....))))....((.(((((....))))).))...))))).))))..(.....)))---))))... ( -31.30) >consensus ____________CUGGCCUUUGCCAAUUAUUGCAAUGCGCUCUGCGCACGCACCUCCGGCAAUUAAAAUUAAUUUC_CUCGCAGACGCCAUUGUCUGGCGCAAAAGCCAGG ............(((((..((((........)))).......(((((..((.......)).....................((((((....)))))))))))...))))). (-19.23 = -20.03 + 0.81)

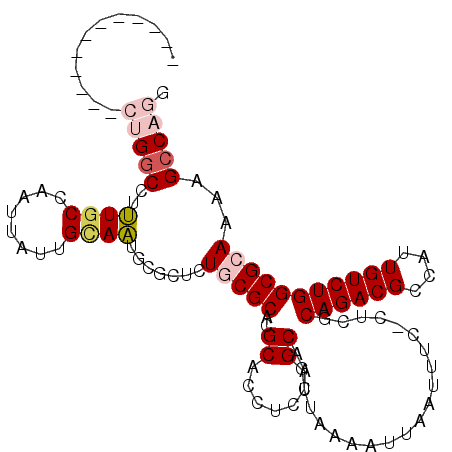
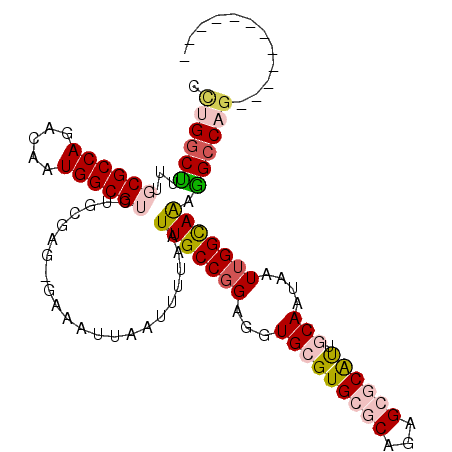
| Location | 11,467,439 – 11,467,537 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -27.04 |
| Energy contribution | -28.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11467439 98 - 22407834 UCUGGCUUUUGCGCCAGACAAUGGCGUCUGCGAG-GAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCGCAGAGCGCAUUGCAAUAAUUGGUAAAGGCCAG------------ .(((((((((((((((.....)))).(((((((.-.(((.....)))..))).))))..(((((((((...))))).)))).......)))))))))))------------ ( -38.10) >DroPse_CAF1 31611 101 - 1 CUGGGCC---CCGCCAGACAAUGGCGUCUGCGAGGGAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCG-------CACUACAAUAAUUGGCAGAGGCCAGAUGCAGAGGCCA ...((((---.(((((.....)))))((((((..((.(((((....))))).)).(.(((((....)-------)))).).....(((((....))))).)))))))))). ( -38.60) >DroEre_CAF1 31503 98 - 1 ACUGGCUUUUGCGCCAGACAAUGGCGUCUGCGAG-GAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCGCAGAGCGCGUUGCAAUAAUUGGCAAAGGCCAG------------ .(((((((((((((((.....))))...((((.(-(.(((((....))))).))......((((((.....)))))))))).......)))))))))))------------ ( -39.60) >DroYak_CAF1 31891 98 - 1 UCUGGCUUUUGCGCCAGACAAUGGCGUCUGCGAG-GAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCGCAGAGCGCAUUGCAAUAAUUGGCAAAGGCCAG------------ .(((((((((((((((.....)))).(((((((.-.(((.....)))..))).))))..(((((((((...))))).)))).......)))))))))))------------ ( -40.10) >DroAna_CAF1 29473 102 - 1 CCUGUCUCUUGCGCCAGACAAUGGCGUCUGCUAG-GAAAUUAAUUUUAAUUGCCCGAGGUGCGUGCGCAAUGCGCAUUGCAAUAAUUGGCAAAAGCCAGAUCU-------- ....((((..((((((.....))))))......(-(.(((((....)))))..))))))(((((((((...))))).))))....(((((....)))))....-------- ( -31.40) >DroPer_CAF1 32616 95 - 1 CCGGGCC---CCGCCAGACAAUGGCGUCUGCGAGGGAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCG-------CACUACAAUAAUUGGCAGAGGCCA------GAGGCCA ((.((((---.(((((.....)))))(((((.((((.(((((....))))).)).(.(((((....)-------)))).).....)).))))))))).------..))... ( -34.90) >consensus CCUGGCUUUUGCGCCAGACAAUGGCGUCUGCGAG_GAAAUUAAUUUUAAUUGCCGGAGGUGCGUGCGCAGAGCGCAUUGCAAUAAUUGGCAAAGGCCAG____________ .((((((...((((((.....))))))......................(((((((...(((((((((...)))))).)))....))))))).))))))............ (-27.04 = -28.10 + 1.06)

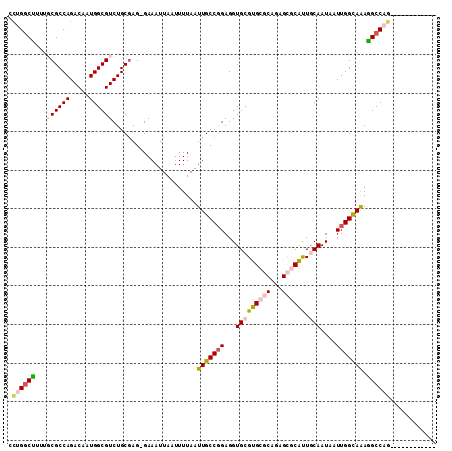
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:01 2006