| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,465,534 – 11,465,644 |
| Length | 110 |
| Max. P | 0.620452 |

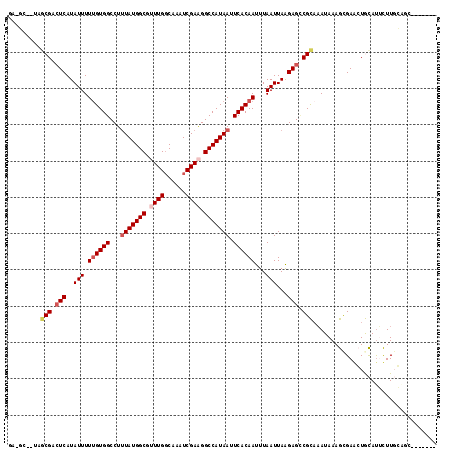
| Location | 11,465,534 – 11,465,644 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
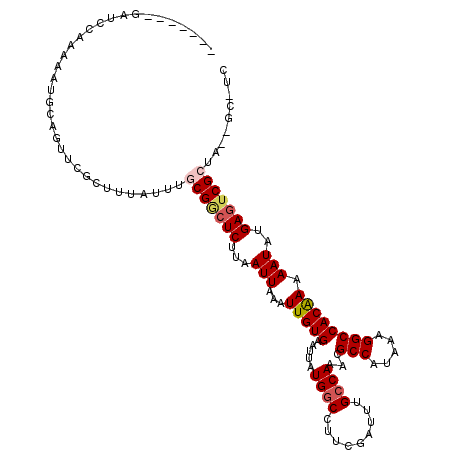
>2L_DroMel_CAF1 11465534 110 + 22407834 -AUCGCAUCGCGACUCAUAUUUUUGUGGCCUUUAUGGCGAUUGGCAAAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGCCGCAAAUAAAGCGAGCUGCAUUCUUGGCCC------- -.((((...(((.(((..(((.((((((...(((((((..((((....))))..))))))).))))))...)))...))).))).......))))(((........)))..------- ( -27.20) >DroVir_CAF1 76686 106 + 1 GA-GC---AGCGAGUCAUAUUUUUGUGGCCUUUAUGGCGUUUGGCAAAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGGCGUAAAUAAAGCGAACUAUAUGUUUCCAU-------- ((-((---(...((((......((((((...(((((((.(((((....))))).))))))).))))))............((.........))).)))...)))))....-------- ( -24.50) >DroGri_CAF1 107202 116 + 1 GG-GCAAUAGCGACUCAUAUUUUUGUGGCCU-UAUGGCGUUUGCCAGAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGUCGCCAAUAAAGUGAAGUGCACUUUUGCUGUCUCACAG .(-(((...(((((((..(((.((((((..(-((((((.((((......)))).))))))).))))))...)))...)))))))....((((((.....))))))))))......... ( -37.60) >DroEre_CAF1 29599 103 + 1 G--------GCGACUCAUAUUUUUGUGGCCUUUAUGGCGAUUGGCAAAUCGAGGGCCAUAAUUCACAAUUUAAUUAGGAGCCGCAAAUAAAGCGAGCUGCAUUCUUGGAAC------- .--------(((.(((.....(((((((((((((((((..((((....))))..)))))))...............)).))))))))......))).)))...........------- ( -26.86) >DroMoj_CAF1 60145 111 + 1 CA-GC--UAUCGACUCAUAUUUUCGUGGCCUUUAUGGCGUUUGGCAAAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGCCGUAUAUAAAGCGAAGUACAUGUUUUCAUCAUC---- ..-((--(((((.(((..(((...((((...(((((((.(((((....))))).))))))).)))).....)))...))).)).))....)))((((.......))))......---- ( -21.60) >DroAna_CAF1 28151 110 + 1 -GUCACUGAGCGACUCAUAUUUUUGUGGCCUUUAUGGCGUUUGGCAAAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGCCGCAAAUAAAGCGAACUGCAUUCUUGGGGG------- -........(((.(((..(((.((((((...(((((((.(((((....))))).))))))).))))))...)))...))).)))............((.((....)).)).------- ( -29.40) >consensus GA_GC__UAGCGACUCAUAUUUUUGUGGCCUUUAUGGCGUUUGGCAAAUCGAAGGCCAUAAUUCACAAUUUAAUUAAGAGCCGCAAAUAAAGCGAACUGCAUUCUUGCAGC_______ .........(((.(((..(((.((((((...(((((((.((((......)))).))))))).))))))...)))...))).))).................................. (-22.21 = -23.10 + 0.89)


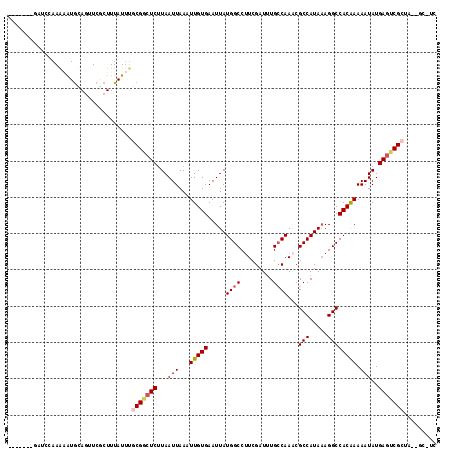
| Location | 11,465,534 – 11,465,644 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11465534 110 - 22407834 -------GGGCCAAGAAUGCAGCUCGCUUUAUUUGCGGCUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUUUGCCAAUCGCCAUAAAGGCCACAAAAAUAUGAGUCGCGAUGCGAU- -------((((..........))))((.....(((((((((.(((((........)))))((((((((((((....))))).....))))))).........))))))))).))...- ( -30.60) >DroVir_CAF1 76686 106 - 1 --------AUGGAAACAUAUAGUUCGCUUUAUUUACGCCUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUUUGCCAAACGCCAUAAAGGCCACAAAAAUAUGACUCGCU---GC-UC --------(((....)))(((((((((...(((((...........))))).)))))))))((((((......((.....))....))))))..................---..-.. ( -20.20) >DroGri_CAF1 107202 116 - 1 CUGUGAGACAGCAAAAGUGCACUUCACUUUAUUGGCGACUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUCUGGCAAACGCCAUA-AGGCCACAAAAAUAUGAGUCGCUAUUGC-CC ..........(((((((((.....))))))..(((((((((.(((((........)))))(((((((.....((((....)))).)-)))))).........))))))))).)))-.. ( -35.10) >DroEre_CAF1 29599 103 - 1 -------GUUCCAAGAAUGCAGCUCGCUUUAUUUGCGGCUCCUAAUUAAAUUGUGAAUUAUGGCCCUCGAUUUGCCAAUCGCCAUAAAGGCCACAAAAAUAUGAGUCGC--------C -------.......(((((.((....)).)))))(((((((.((.((...(((((.....((((.........))))...(((.....)))))))))).)).)))))))--------. ( -23.90) >DroMoj_CAF1 60145 111 - 1 ----GAUGAUGAAAACAUGUACUUCGCUUUAUAUACGGCUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUUUGCCAAACGCCAUAAAGGCCACGAAAAUAUGAGUCGAUA--GC-UG ----.....................(((....((.((((((...(((...(((((.....((((.........))))...(((.....)))))))).)))..)))))))))--))-.. ( -19.90) >DroAna_CAF1 28151 110 - 1 -------CCCCCAAGAAUGCAGUUCGCUUUAUUUGCGGCUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUUUGCCAAACGCCAUAAAGGCCACAAAAAUAUGAGUCGCUCAGUGAC- -------................(((((......(((((((...(((...(((((.....((((.........))))...(((.....)))))))).)))..)))))))..))))).- ( -25.80) >consensus _______GAUCCAAAAAUGCAGUUCGCUUUAUUUGCGGCUCUUAAUUAAAUUGUGAAUUAUGGCCUUCGAUUUGCCAAACGCCAUAAAGGCCACAAAAAUAUGAGUCGCUA__GC_UC ..................................(((((((...(((...(((((.....((((.........))))...(((.....)))))))).)))..)))))))......... (-18.10 = -18.68 + 0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:59 2006