
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,464,258 – 11,464,446 |
| Length | 188 |
| Max. P | 0.863204 |

| Location | 11,464,258 – 11,464,356 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -21.90 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
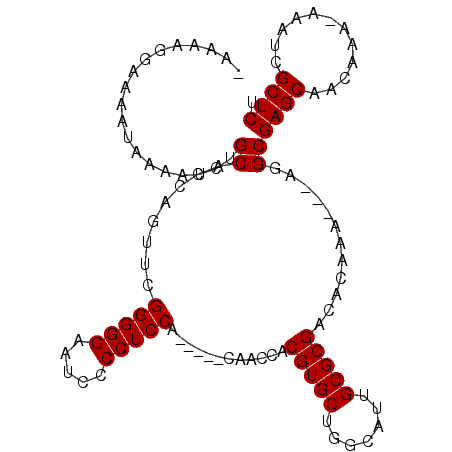
>2L_DroMel_CAF1 11464258 98 + 22407834 -AAAAGGAAAAUAAAAUCUGCACCAGUUCGGGGGAAUCCCCUCCA-----CAACCACGUGCUGGCAUUGCGCGACACAAA---AGGCGAGCAACAAA-AAAUCGCUCU -..................((........(((((.....))))).-----......(((((.......))))).......---..))((((......-.....)))). ( -22.00) >DroSec_CAF1 27816 101 + 1 -AAAAGGAAAAUAAAAUCUGCACCAGUUCGGGGGAAUCCCCUCCA-----CAACCACGUGCUGGCAUUGCGCGACACAAACACAGGCGAGCAACAAA-AAAUCGCUCU -...............((((.........(((((.....))))).-----......(((((.......))))).........)))).((((......-.....)))). ( -22.10) >DroSim_CAF1 30128 102 + 1 -AAAAGGAAAAUAAAAUCUGCACCAGUUCGGGGGAAUCCCCUCCA-----CAACCACGUGCUGGCAUUGCGCGACACAAACACAGGCGAGCAACAAAAAAAUCGCUCU -...............((((.........(((((.....))))).-----......(((((.......))))).........)))).((((............)))). ( -22.00) >DroEre_CAF1 28146 99 + 1 GAAAAGGGAAAUAAAAUCUGCACCGGUUCGGGGGAAUCCCCUCCA-----CAACCACGUGCUGGCAUUGCGCGACACAAA---AGGCGAGCAACAAA-AAAUCGCUCU ...................((...((((.(((((.....))))).-----.)))).(((((.......))))).......---..))((((......-.....)))). ( -25.40) >DroYak_CAF1 28658 104 + 1 GAAAAGGAAAAUAAAAUCUGCACCAGUUCGGGGGAAUCCCCUCCAACAACCAACCACGUGCUGGCAUUGCGCGACACAAA---AGGCGAGCAACAAA-AAAUCGCUCU ...................((....(((.(((((....))))).))).........(((((.......))))).......---..))((((......-.....)))). ( -23.90) >consensus _AAAAGGAAAAUAAAAUCUGCACCAGUUCGGGGGAAUCCCCUCCA_____CAACCACGUGCUGGCAUUGCGCGACACAAA___AGGCGAGCAACAAA_AAAUCGCUCU ...................((........(((((.....)))))............(((((.......)))))............))((((............)))). (-21.90 = -21.90 + 0.00)


| Location | 11,464,258 – 11,464,356 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.57 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
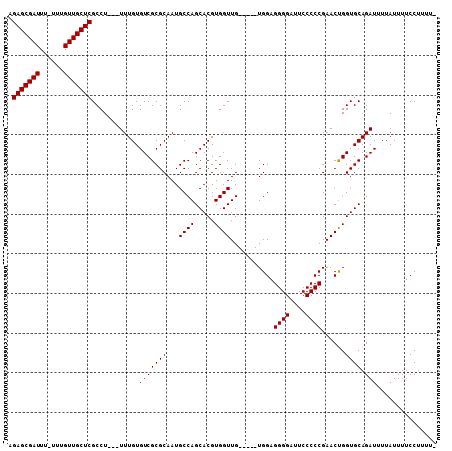
>2L_DroMel_CAF1 11464258 98 - 22407834 AGAGCGAUUU-UUUGUUGCUCGCCU---UUUGUGUCGCGCAAUGCCAGCACGUGGUUG-----UGGAGGGGAUUCCCCCGAACUGGUGCAGAUUUUAUUUUCCUUUU- .(((((((..-...)))))))....---((((..((.((((..((((.....))))))-----))..((((....)))).....))..))))...............- ( -29.40) >DroSec_CAF1 27816 101 - 1 AGAGCGAUUU-UUUGUUGCUCGCCUGUGUUUGUGUCGCGCAAUGCCAGCACGUGGUUG-----UGGAGGGGAUUCCCCCGAACUGGUGCAGAUUUUAUUUUCCUUUU- .(((((((..-...)))))))..((((..(((((...))))).(((((((((....))-----))..((((....))))...)))))))))................- ( -31.60) >DroSim_CAF1 30128 102 - 1 AGAGCGAUUUUUUUGUUGCUCGCCUGUGUUUGUGUCGCGCAAUGCCAGCACGUGGUUG-----UGGAGGGGAUUCCCCCGAACUGGUGCAGAUUUUAUUUUCCUUUU- .(((((((......)))))))..((((..(((((...))))).(((((((((....))-----))..((((....))))...)))))))))................- ( -31.80) >DroEre_CAF1 28146 99 - 1 AGAGCGAUUU-UUUGUUGCUCGCCU---UUUGUGUCGCGCAAUGCCAGCACGUGGUUG-----UGGAGGGGAUUCCCCCGAACCGGUGCAGAUUUUAUUUCCCUUUUC .(((((((..-...)))))))((..---.(((((...))))).))..((((.(((((.-----(((.(((....)))))))))))))))................... ( -31.70) >DroYak_CAF1 28658 104 - 1 AGAGCGAUUU-UUUGUUGCUCGCCU---UUUGUGUCGCGCAAUGCCAGCACGUGGUUGGUUGUUGGAGGGGAUUCCCCCGAACUGGUGCAGAUUUUAUUUUCCUUUUC .(((((((..-...)))))))....---((((..((...((((((((((.....))))).)))))..((((....)))).....))..))))................ ( -31.80) >consensus AGAGCGAUUU_UUUGUUGCUCGCCU___UUUGUGUCGCGCAAUGCCAGCACGUGGUUG_____UGGAGGGGAUUCCCCCGAACUGGUGCAGAUUUUAUUUUCCUUUU_ .(((((((......)))))))............(((((((...((((.....))))...........((((....))))......)))).)))............... (-25.90 = -25.90 + 0.00)


| Location | 11,464,356 – 11,464,446 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -19.38 |
| Energy contribution | -20.44 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
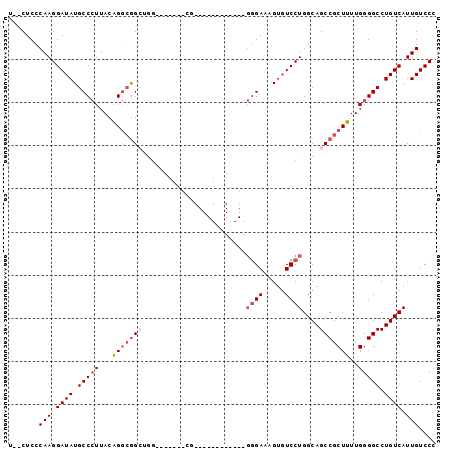
>2L_DroMel_CAF1 11464356 90 - 22407834 U--CUCCCAAGGAUAUGCCCUUACAGGAGGCUGG-------CGGGGGUAACGGCGGGGAAAGUGUCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC .--....(((.((((.(((((...(((.(((((.-------(((((...((..........)).))))).))))).)))..))))).)))).))).... ( -34.60) >DroPse_CAF1 28638 77 - 1 UCUCUCCCAAGGAUAUGCCGUUACGG-CAGCAGC-------A--------------AGAGCGUGUCCUGGCAGCCGCUCUUGGGGCCUGUCAUUGUCCC ..........((((((((((...)))-))((..(-------(--------------(((((((((....)))..))))))))..)).......))))). ( -28.10) >DroSec_CAF1 27917 76 - 1 U--CUCCCAAGGAUAUGCCCUUACAGGCGGCUGG-------CG------------GGGAAAG--UCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC .--....(((.((((.(((((...(((((((((.-------((------------(((....--))))).)))))))))..))))).)))).))).... ( -40.10) >DroSim_CAF1 30230 78 - 1 U--CUCCCAAGGAUAUGCCCUUACAGGCGGCUGG-------CG------------GGGAAAGUGUCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC .--....(((.((((.(((((...(((((((((.-------((------------(((......))))).)))))))))..))))).)))).))).... ( -40.30) >DroEre_CAF1 28245 75 - 1 ---CUCCCAAGGAUAUGCCCUUACAGGCGGGCAG---------------------GGGAAAGUGUCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC ---....(((.((((.(((((...((((((((..---------------------((((.....)))).))..))))))..))))).)))).))).... ( -28.90) >DroYak_CAF1 28762 97 - 1 U--CUCCCAAGGAUAUGCCCUUACAGGCGGGAAGAGGGGAGAGGGGGUAACGGCGGGGAAAGUGUCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC (--(((((.......((((......)))).......)))))).(((..((.((((((((.....))).....(((.(....).)))))))).))..))) ( -36.64) >consensus U__CUCCCAAGGAUAUGCCCUUACAGGCGGCUGG_______CG____________GGGAAAGUGUCCUGGCAGCCGCUUUUGGGGCCUGUCAUUGUCCC .......(((.((((.(((((...(((((((........................((((.....))))....)))))))..))))).)))).))).... (-19.38 = -20.44 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:57 2006