
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,438,261 – 11,438,394 |
| Length | 133 |
| Max. P | 0.914473 |

| Location | 11,438,261 – 11,438,369 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
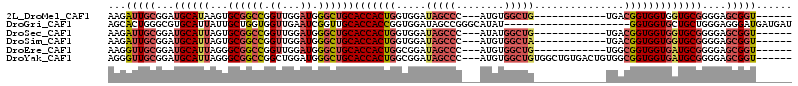
>2L_DroMel_CAF1 11438261 108 - 22407834 ---------CAGCCACAU---GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUUAUGCAUCCGCAAUCUUCGCCGGGCUCGCAAUCCCACCUGGAUCACCUGCCCACACC ---------........(---((((.(((((...((((((((((((........((..(((....)))..))((.......)).))))).)))...)))).))))).....))))).... ( -35.00) >DroVir_CAF1 50071 105 - 1 ---------------CAUGCCCGCCUAUCCGCCUGUGGUGCAGCCCAUCCAACCACCAGCAAUCAUGCACGCGCAGUGCUCACCUGGAUCGCAGACCCAUCUGGAUCAGUUGCCGACUCC ---------------..(((((((..........)))).)))................(((((...((((.....)))).......((((.((((....)))))))).)))))....... ( -24.90) >DroGri_CAF1 72276 106 - 1 --------------AUAUGCCCGGCUAUCCACCGGUGGUGCAACCGAUUCAACCACCAGCAAUAAUGCACGCCCAGUGCUCGCCCGGCUCCCAUUCGCAUCUAGAUCAGCUGCCGACACC --------------....(((.(((........((((((............)))))).........((((.....))))..))).)))......(((((.((.....)).)).))).... ( -28.80) >DroSim_CAF1 2050 108 - 1 ---------UAGCCACAU---GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUAAUGCAUCCGCAAUCUUCGCCCGGUUCCCAAUCCCACCUAGAUCACCUGCCCACGCC ---------(((((....---.))))).......((((.((((.......((((((.((......(((....))).....)).))))))....(((.......)))...))))))))... ( -28.80) >DroEre_CAF1 2055 108 - 1 ---------CAGCCACAU---GGGCUAUCCGCCAGUGGUGCAGCCCAUCCAACCGGCCGCCCUAAUGCAUCCGCAACCUUCGCCGGGCUCCCAAUCCCACCUGGAUCACCUGCCCACGCC ---------.((((....---.))))........((((.((((...(((((...(((........(((....)))......)))(((........)))...)))))...))))))))... ( -32.74) >DroYak_CAF1 2056 117 - 1 UCACAGCCACAGCCACAU---GGGCUAUCCGCCAGUGGUGCAGCCCAUCCAGCCGGCCGCCCUAAUGCAUCCGCAACCCUCGCCGGGCUCCCAAUCCCACCUGGAUCACCUGCCCACGCC ....((((.((......)---)))))........((((.((((...((((((..(((........(((....)))......)))(((........)))..))))))...))))))))... ( -36.64) >consensus _________CAGCCACAU___GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUAAUGCAUCCGCAAUCCUCGCCGGGCUCCCAAUCCCACCUGGAUCACCUGCCCACGCC .....................(((((..(((....)))...))))).......(((..((......))..)))...........((((.....((((.....)))).....))))..... (-17.41 = -17.92 + 0.50)


| Location | 11,438,301 – 11,438,394 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -25.56 |
| Energy contribution | -27.15 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
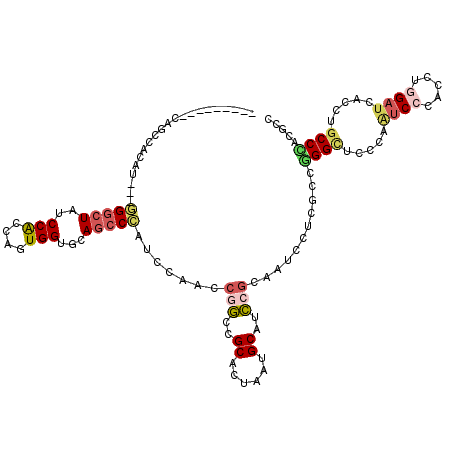
>2L_DroMel_CAF1 11438301 93 + 22407834 AAGAUUGCGGAUGCAUAAGUGCGGCCGGUUGGAUGGGCUGCACCACUGGUGGAUAGCCC---AUGUGGCUG------------UGACGGUGGUGGUGCGGGGAGCGGU------ ...(((((...(((((..((((((((.((...)).))))))))(((((.((.((((((.---....)))))------------)..)).))))))))))....)))))------ ( -39.10) >DroGri_CAF1 72316 93 + 1 AGCACUGGGCGUGCAUUAUUGCUGGUGGUUGAAUCGGUUGCACCACCGGUGGAUAGCCGGGCAUAU---------------------GGUGGUGCUGCUGGGAGGGAUGAUGAU .((((.....))))((((((.(((((......)))(((.(((((.(((((.....)))))((....---------------------.))))))).)))...)).))))))... ( -30.90) >DroSec_CAF1 2091 93 + 1 AAGAUUGCGGAUGCAUUAGUGCGGCCGGUUGGAUGGGCUGCACCACUGGUGGAUAGCCC---AUAUGGCUG------------UGACGGUGGUGGUGCGGGGAGCGGU------ ...(((((...(((((((((((((((.((...)).))))))))(((((....((((((.---....)))))------------)..))))).)))))))....)))))------ ( -39.40) >DroSim_CAF1 2090 93 + 1 AAGAUUGCGGAUGCAUUAGUGCGGCCGGUUGGAUGGGCUGCACCACUGGUGGAUAGCCC---AUGUGGCUA------------UGACGGUGGUGGUGCGGGGAGCGGU------ ...(((((...(((((((((((((((.((...)).))))))))(((((....((((((.---....)))))------------)..))))).)))))))....)))))------ ( -39.40) >DroEre_CAF1 2095 93 + 1 AAGGUUGCGGAUGCAUUAGGGCGGCCGGUUGGAUGGGCUGCACCACUGGCGGAUAGCCC---AUGUGGCUG------------UGGCGGUGGUGAUGCGGGGAGCGGU------ ....((((...((((((...((((((.((...)).))))))(((((((.(..((((((.---....)))))------------)).)))))))))))))....)))).------ ( -40.00) >DroYak_CAF1 2096 105 + 1 AGGGUUGCGGAUGCAUUAGGGCGGCCGGCUGGAUGGGCUGCACCACUGGCGGAUAGCCC---AUGUGGCUGUGGCUGUGACUGUGGCGGUGGUGAUGCGGGGAGCGGU------ .(.((..(.(.(((..(((.(((((((((((.((((((((..((......)).))))))---)).)))))..))))))..)))..))).).)..)).)..........------ ( -41.90) >consensus AAGAUUGCGGAUGCAUUAGUGCGGCCGGUUGGAUGGGCUGCACCACUGGUGGAUAGCCC___AUGUGGCUG____________UGACGGUGGUGGUGCGGGGAGCGGU______ ...(((((...((((((...((((((.((...)).))))))(((((((.....(((((........)))))...............)))))))))))))....)))))...... (-25.56 = -27.15 + 1.59)

| Location | 11,438,301 – 11,438,394 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -15.73 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11438301 93 - 22407834 ------ACCGCUCCCCGCACCACCACCGUCA------------CAGCCACAU---GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUUAUGCAUCCGCAAUCUU ------...((.....(((((((........------------.((((....---.))))........))))))).(((.........))).)).....(((....)))..... ( -24.13) >DroGri_CAF1 72316 93 - 1 AUCAUCAUCCCUCCCAGCAGCACCACC---------------------AUAUGCCCGGCUAUCCACCGGUGGUGCAACCGAUUCAACCACCAGCAAUAAUGCACGCCCAGUGCU ...............(((((((.....---------------------...)))..(((........((((((............)))))).(((....)))..)))...)))) ( -20.90) >DroSec_CAF1 2091 93 - 1 ------ACCGCUCCCCGCACCACCACCGUCA------------CAGCCAUAU---GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUAAUGCAUCCGCAAUCUU ------...((.....(((((((........------------.((((....---.))))........))))))).(((.........))).)).....(((....)))..... ( -24.13) >DroSim_CAF1 2090 93 - 1 ------ACCGCUCCCCGCACCACCACCGUCA------------UAGCCACAU---GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUAAUGCAUCCGCAAUCUU ------...((.....(((((((.......(------------(((((....---.))))))......))))))).(((.........))).)).....(((....)))..... ( -26.82) >DroEre_CAF1 2095 93 - 1 ------ACCGCUCCCCGCAUCACCACCGCCA------------CAGCCACAU---GGGCUAUCCGCCAGUGGUGCAGCCCAUCCAACCGGCCGCCCUAAUGCAUCCGCAACCUU ------...((.....((((.(((((.((..------------.((((....---.))))....))..)))))((.(((.........))).))....))))....))...... ( -22.10) >DroYak_CAF1 2096 105 - 1 ------ACCGCUCCCCGCAUCACCACCGCCACAGUCACAGCCACAGCCACAU---GGGCUAUCCGCCAGUGGUGCAGCCCAUCCAGCCGGCCGCCCUAAUGCAUCCGCAACCCU ------...((.....((((.......(((...((.......)).((...((---(((((..(((....)))...)))))))...)).))).......))))....))...... ( -23.24) >consensus ______ACCGCUCCCCGCACCACCACCGUCA____________CAGCCACAU___GGGCUAUCCACCAGUGGUGCAGCCCAUCCAACCGGCCGCACUAAUGCAUCCGCAAUCUU ................((.....................................(((((..(((....)))...)))))........((..((......))..))))...... (-15.73 = -15.45 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:49 2006