| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,436,644 – 11,436,735 |
| Length | 91 |
| Max. P | 0.644105 |

| Location | 11,436,644 – 11,436,735 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -22.57 |
| Energy contribution | -24.35 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
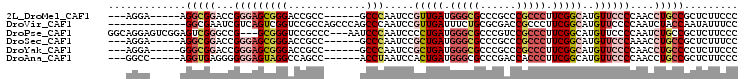
>2L_DroMel_CAF1 11436644 91 - 22407834 ---AGGA-----AGGCGGACCGGGAGCGGGACCGCC------GCCCAAUCCGUUGAUGGGCGCCCGCCCGCCCUUCGGCAUGUUCCCCAACCUGCCGCUCUUCCC ---.(((-----((((((...(((((((((.(.(.(------(((((.((....)))))))).).))))(((....)))..)))))).......)))).))))). ( -44.90) >DroVir_CAF1 48454 92 - 1 -------------GGCGAAUCGUCAGUCGGUCCGCCAGCCCAGCCCAAUCCGUUGAUUUCUGCGCGACCGCCCUUCGGCAUGUUCCCCAAUCUACCAAUAUUUCC -------------((.((((........(((((((.((..((((.......))))....))))).))))(((....)))..)))).))................. ( -21.20) >DroPse_CAF1 469 99 - 1 GGCAGGAGUCGGAGUCGGGCCG---GCGGGUCCGCCC---AAUCCCAAUCCCCUGAUGGGCGCCCGUCCGCCCUUCGGCAUGUUCCCCAAUCUGCCGCUCUUCCC ((((((.((((((...((((.(---(((((..(((((---(.((..........)))))))))))))).)))))))))).((.....)).))))))......... ( -43.90) >DroSec_CAF1 434 91 - 1 ---AGGA-----AGGCGGACCGGGAGCGGGACCGCC------GCCCAAUCCGCUGAUGGGCGCCCGCCCGCCCUUCGGCAUGUUCCCAAACCUGCCGCUCUUUCC ---.(((-----((((((...(((((((((......------.))).....(((((.(((((......))))).)))))..)))))).......)))).))))). ( -43.10) >DroYak_CAF1 439 91 - 1 ---AGGA-----GGGCGGACCGGGAGCGGGACCGCC------GCCCAAUCCGCUGAUGGGCGCCCGCCCGCCCUUCGGCAUGUUCCCCAACCUGCCCCUCUUCCC ---.(((-----((((((..((((.(((....)))(------(((((.((....)))))))))))).)))))))))((((.(((....))).))))......... ( -46.60) >DroAna_CAF1 438 91 - 1 ---GGCC-----AGGUGAGGGGGGAGUAGGCCAGCC------ACCUAAUCCACUGAUGGGCGCCCGACCACCCUUCGGCAUGUUCCCCAACCUGCCGCUCUUCCC ---(((.-----((((..(((((((.((((......------.)))).))).((((.(((.(......).))).))))......)))).)))))))......... ( -34.80) >consensus ___AGGA_____AGGCGGACCGGGAGCGGGACCGCC______GCCCAAUCCGCUGAUGGGCGCCCGCCCGCCCUUCGGCAUGUUCCCCAACCUGCCGCUCUUCCC .............(((((...(((((((((.............))).....(((((.(((((......))))).)))))..))))))....)))))......... (-22.57 = -24.35 + 1.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:47 2006