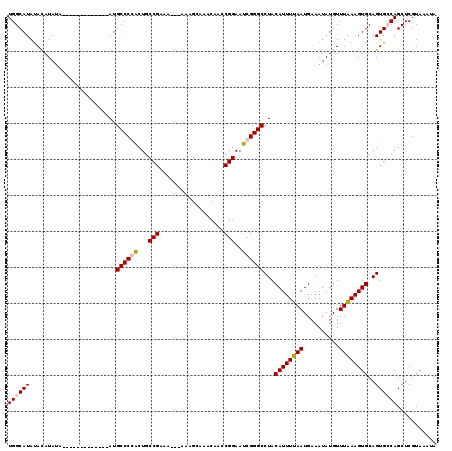
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,208,836 – 1,209,021 |
| Length | 185 |
| Max. P | 0.908698 |

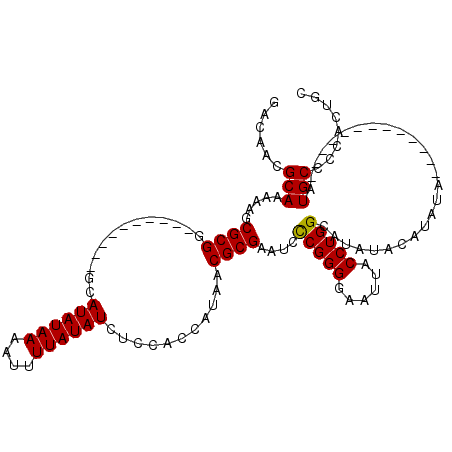
| Location | 1,208,836 – 1,208,945 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1208836 109 + 22407834 GACAACGCAAAAAGCGCGG----------GCAUAUAAAAUUUUAUAUCUUCCCCAUAACGCGAAUCUCGGGGAAUUACCUGGCAUAUACAUAUAUAUAUAUUGUAUAAUGCCCGACUGC .....(((.....)))(((----------((((...............((((((..............))))))..........(((((((((....))).)))))))))))))..... ( -26.64) >DroSim_CAF1 4953 96 + 1 GACAACGCAAAAAGCGCGG----------GCAUAUAAAAUUUUAUAUCUCCACCAUAACGCGAAUCUCGGGGAAUUACCUGGCAUAUAUAUAUA-------------AUGCCCGACUGC .....(((.....)))(((----------((((((((....)))))....................(((((......)))))............-------------.))))))..... ( -19.30) >DroEre_CAF1 4285 96 + 1 GACAACGCAAAAAGCGCGG----------GCAUAUAAAAUUUUAUAUCUCCAGCAUAACGCGAAUCCCGGGGAAUUACCUGGCAUACAUAUAAA-------------AUGCCCCGCUGC ............((((.((----------((((.......(((((((.....((.....))(.((.(((((......))))).)).))))))))-------------)))))))))).. ( -26.61) >DroYak_CAF1 4657 106 + 1 GACAACGCAAAAAGCGCGGGCAGAAGCGGGCAUAUAAAAUUUUAUAUCUGCACCAUAACGCGAAUCCCGGGGAAUUACCUGGCAUAUACAUAUA-------------AUGCCCCGCUGC ............((((.(((((...((((..(((........)))..))))...............(((((......)))))............-------------.))))))))).. ( -31.10) >consensus GACAACGCAAAAAGCGCGG__________GCAUAUAAAAUUUUAUAUCUCCACCAUAACGCGAAUCCCGGGGAAUUACCUGGCAUAUACAUAUA_____________AUGCCCCACUGC ......(((.....((((.............((((((....))))))...........))))....(((((......)))))..........................)))........ (-15.30 = -15.05 + -0.25)

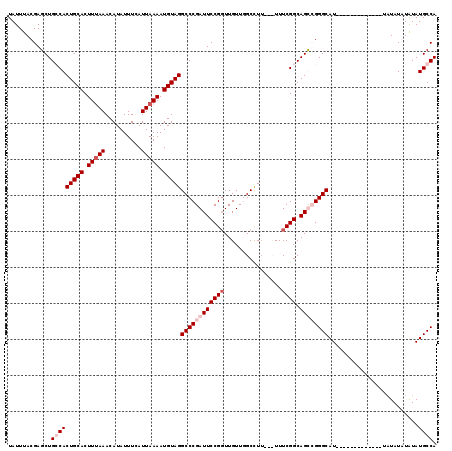
| Location | 1,208,905 – 1,209,021 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -19.53 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1208905 116 + 22407834 UGGCAUAUACAUAUAUAUAUAUUGUAUAAUGCCCGACUGCCGAAA---AAGGCAAACAAGCGGAAUCGGGCCUACAUUUUAAUGAAAUAUGUUUAAAGUGCAGUGCCAGCUCGUAAAUA .(((((((((((((....))).))))).)))))....((((....---..)))).....((((.....(((((.((((((((..........)))))))).)).)))...))))..... ( -29.40) >DroSim_CAF1 5022 104 + 1 UGGCAUAUAUAUAUA-------------AUGCCCGACUGCCGAAAA--AAGGCAAACAACCGGAAUCGGGCCUACAUUUUAAUGAAAUAUGUUUAAAGUGCAGUGCCAGCUCGUAAAUA ((((((.........-------------..((((((.((((.....--..))))..(....)...))))))...((((((((..........))))))))..))))))........... ( -26.60) >DroEre_CAF1 4354 102 + 1 UGGCAUACAUAUAAA-------------AUGCCCCGCUGCCGAAA----AAGCCAACAACCGGAAUCGGGCCUACAUUUUAAUGAAAUAUGUUUGAAGUGCAGUGGCAGCUCGUAAAUA .(((((.........-------------)))))..(((((((...----..........(((....)))..((.((((((((..........)))))))).)))))))))......... ( -25.70) >DroYak_CAF1 4736 106 + 1 UGGCAUAUACAUAUA-------------AUGCCCCGCUGCCGAAAAAAAAAGCCAACAACCGGAAUCGGGCCUACAUUUUAAUGAAAUAUGUUUAAAGUGCAGUGGCAGCUCGUAAAUA .(((((.........-------------)))))..(((((((.................(((....)))..((.((((((((..........)))))))).)))))))))......... ( -25.60) >consensus UGGCAUAUACAUAUA_____________AUGCCCCACUGCCGAAA___AAAGCAAACAACCGGAAUCGGGCCUACAUUUUAAUGAAAUAUGUUUAAAGUGCAGUGCCAGCUCGUAAAUA ((((((........................((((((...(((..................)))..))))))...((((((((..........))))))))..))))))........... (-19.53 = -20.10 + 0.56)

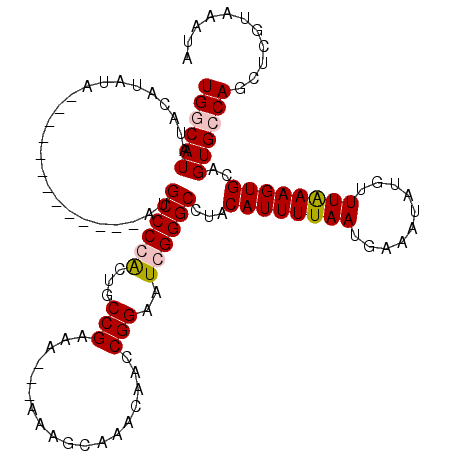
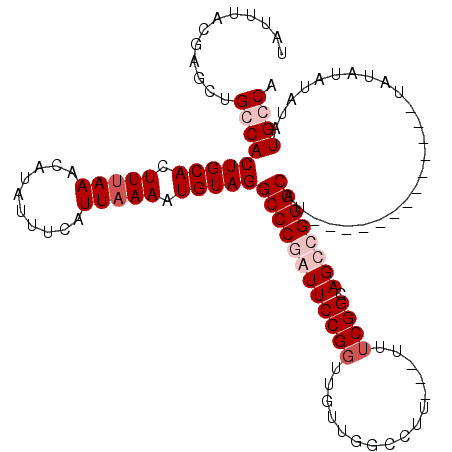
| Location | 1,208,905 – 1,209,021 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -20.54 |
| Energy contribution | -22.54 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1208905 116 - 22407834 UAUUUACGAGCUGGCACUGCACUUUAAACAUAUUUCAUUAAAAUGUAGGCCCGAUUCCGCUUGUUUGCCUU---UUUCGGCAGUCGGGCAUUAUACAAUAUAUAUAUAUGUAUAUGCCA .....((((((.(((.(((((.(((((..........))))).)))))))).(....)))))))(((((..---....)))))...(((((.(((((.(((....))))))))))))). ( -31.90) >DroSim_CAF1 5022 104 - 1 UAUUUACGAGCUGGCACUGCACUUUAAACAUAUUUCAUUAAAAUGUAGGCCCGAUUCCGGUUGUUUGCCUU--UUUUCGGCAGUCGGGCAU-------------UAUAUAUAUAUGCCA ............(((((((((.(((((..........))))).)))))((((((((((((..(........--)..)))).))))))))..-------------..........)))). ( -30.80) >DroEre_CAF1 4354 102 - 1 UAUUUACGAGCUGCCACUGCACUUCAAACAUAUUUCAUUAAAAUGUAGGCCCGAUUCCGGUUGUUGGCUU----UUUCGGCAGCGGGGCAU-------------UUUAUAUGUAUGCCA .........((((((.(((((......................)))))(((((((....))))..)))..----....))))))..(((((-------------.........))))). ( -26.05) >DroYak_CAF1 4736 106 - 1 UAUUUACGAGCUGCCACUGCACUUUAAACAUAUUUCAUUAAAAUGUAGGCCCGAUUCCGGUUGUUGGCUUUUUUUUUCGGCAGCGGGGCAU-------------UAUAUGUAUAUGCCA .........((((((.(((((.(((((..........))))).)))))(((((((....))))..)))..........))))))..(((((-------------.........))))). ( -29.40) >consensus UAUUUACGAGCUGCCACUGCACUUUAAACAUAUUUCAUUAAAAUGUAGGCCCGAUUCCGGUUGUUGGCCUU___UUUCGGCAGCCGGGCAU_____________UAUAUAUAUAUGCCA ............(((((((((.(((((..........))))).)))))((((((((((((................)))).)))))))).........................)))). (-20.54 = -22.54 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:50 2006