| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,432,745 – 11,432,847 |
| Length | 102 |
| Max. P | 0.705029 |

| Location | 11,432,745 – 11,432,847 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -18.41 |
| Energy contribution | -21.36 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
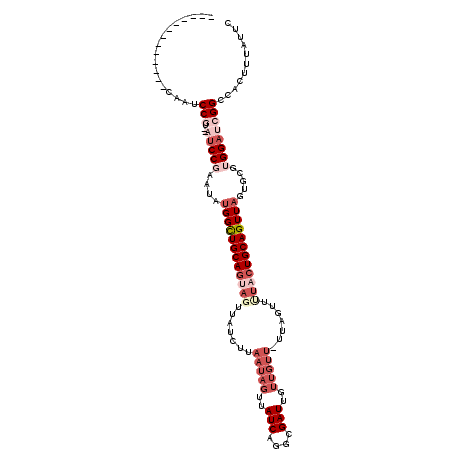
>2L_DroMel_CAF1 11432745 102 + 22407834 -------------CAAUCCGU--AUCCGAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGGCGAUUGUUGUU-UUAGUUUUACUGCAGUUAGUGCGUGGAUCGGCCACUUUAUUC -------------....(((.--(((((....((((((((((((....((.(((((..(((....)))..)))))-..))..)))))))))))).....))))))))........... ( -29.90) >DroPse_CAF1 181989 85 + 1 -------------CAAUCCCA--GCCCCAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGACGAUUG--------GGUUGUUCUGCAGUUUGUGCGUGGAGCGGC---------- -------------.....((.--((.(((....((((((((....((.((((((.(((....))).))))--------)).))..))))))))......))).)))).---------- ( -23.40) >DroSim_CAF1 158058 104 + 1 -------------CAAUCCGUAUAUCCGAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGGCGAUUGUUGUU-UUAGUUUUACUGCAGUUAGUGCGUGGAUCGGCCACUUUAUUC -------------....(((...(((((....((((((((((((....((.(((((..(((....)))..)))))-..))..)))))))))))).....))))))))........... ( -27.60) >DroEre_CAF1 158124 102 + 1 -------------GUAUCCGU--AUCCCAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGGCGAUUGUUGUU-UUAGUUCUACUGCAGUUAGUGCGUGGAUCGGCCACUUUAUUC -------------((..(((.--(((((....((((((((((((....((.(((((..(((....)))..)))))-..))..))))))))))))....).)))))))..))....... ( -31.40) >DroYak_CAF1 156266 116 + 1 CAGAACCCAAUUCCAAACCGU--AUCCGAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGGCGAUUGUUGUUUUUAGUUUUACUGCAGUUAGUGCGUGGAUCGGCCACUUUAUUC .................(((.--(((((....((((((((((((.......(((((..(((....)))..))))).......)))))))))))).....))))))))........... ( -30.34) >DroAna_CAF1 135848 95 + 1 -------------CAUACCCG--AUCCGAAUAUGGUUGCAAUAGUUAUCUUAAUAGUUAUCAGCGGAUUGUUGUU-CUCG-----GUGCAGUUAGUGCGUGGAGCGGCCACUUUAU-- -------------.......(--(((((....((((.((...((....)).....)).)))).))))))((((((-((..-----(..(.....)..)..))))))))........-- ( -21.20) >consensus _____________CAAUCCGU__AUCCGAAUAUGGCUGCAGUAGUUAUCUUAAUAGUUAUCAGGCGAUUGUUGUU_UUAGUUUUACUGCAGUUAGUGCGUGGAUCGGCCACUUUAUUC .................(((...(((((....((((((((((((.......(((((..(((....)))..))))).......)))))))))))).....))))))))........... (-18.41 = -21.36 + 2.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:45 2006