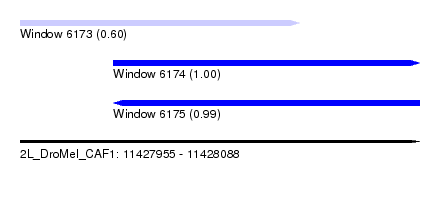
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,427,955 – 11,428,088 |
| Length | 133 |
| Max. P | 0.996244 |

| Location | 11,427,955 – 11,428,048 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -21.65 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
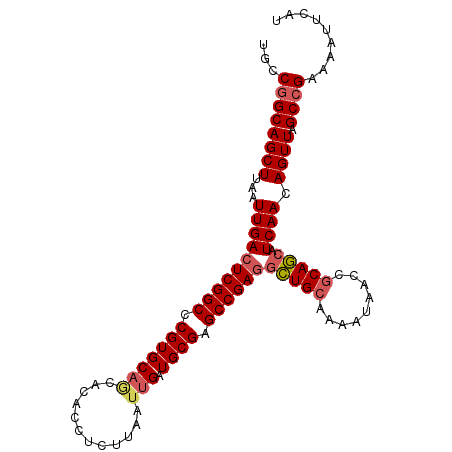
>2L_DroMel_CAF1 11427955 93 + 22407834 ACAAAUGUUUCGCGCUUUUCGGGCCAAUUCAAUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAA ...........(((.......(((((((((...)))))...))))....(((((.(((((((.....))))))).))))).)))......... ( -24.30) >DroSec_CAF1 145489 93 + 1 AGAAAUGUUUCGCGCAGUUCGGGCCAAUUCAAUGCAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAA ...........(((.(((..((((.........(((((..(((....)))..)))))(((((.....)))))))))..))))))......... ( -26.30) >DroSim_CAF1 153022 93 + 1 ACAAAUGUUUCGCGCUGUUCGGGCCAAUUCAAUGCAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAA ...........(((..((..((((.........(((((..(((....)))..)))))(((((.....)))))))))..)).)))......... ( -24.10) >DroEre_CAF1 153107 92 + 1 ACAAAUGUUUCGCGCUGUUCGGGCCAAUUCAAUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUU-UGCAGCCUCGGCUCGCAUCAAUUAA ...........(((.......(((((((((...)))))...))))....(((((.(((((((....)-)))))).))))).)))......... ( -24.30) >DroYak_CAF1 150931 92 + 1 -CAAAUGUUUCGCGCUGUUCGGGCCAAUUCAAUGAAUUUUCGGCUAACUGUUGAUGCUGCAGUUAUUCUGCAGCCUCGGCUCGCAUCAAUUAA -..........(((.......(((((((((...)))))...))))....(((((.(((((((.....))))))).))))).)))......... ( -27.00) >DroAna_CAF1 130965 93 + 1 ACAAAUGUUUCGCACUGUUUGGGCCAAUUCAAUGAAUUUUCGGCUAACUGUUGAUGUUGCGGUUAUUUUCCAAAGUCGGCUCGCACCAAUUAA .....(((...((....(((((((((((((...)))))...)))(((((((.......)))))))....)))))....))..)))........ ( -16.70) >consensus ACAAAUGUUUCGCGCUGUUCGGGCCAAUUCAAUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAA ...................((((((..............(((((.....))))).(((((((.....)))))))...)))))).......... (-21.65 = -21.48 + -0.16)


| Location | 11,427,986 – 11,428,088 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -33.27 |
| Energy contribution | -33.61 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11427986 102 + 22407834 AUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGUGCUGCACGGGCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....(((((.(((((((.....)))))))((((((((...((......)).(((.....))))))))))))))))......))))).. ( -37.30) >DroSec_CAF1 145520 102 + 1 AUGCAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGCGCUGCACGGGCCGAGUCAAUUAAGCUGCAGGCA .(((.....(((((....(((((.(((((((.....)))))))(((((((((((((........)))))(.....))))))))))))))..)))))...))) ( -39.40) >DroSim_CAF1 153053 102 + 1 AUGCAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGUGCUGCACGGGCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....(((((.(((((((.....)))))))((((((((...((......)).(((.....))))))))))))))))......))))).. ( -37.30) >DroEre_CAF1 153138 100 + 1 AUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUU-UGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGUGCUGCACG-GCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....(((((.(((((((....)-))))))(((((((((((((........))))))......)-)))))))))))......))))).. ( -36.60) >DroYak_CAF1 150961 102 + 1 AUGAAUUUUCGGCUAACUGUUGAUGCUGCAGUUAUUCUGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGUGUUGCACGGGCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....(((((.(((((((.....)))))))(((((((((((((........)))))(.....))))))))))))))......))))).. ( -40.20) >DroAna_CAF1 130996 99 + 1 AUGAAUUUUCGGCUAACUGUUGAUGUUGCGGUUAUUUUCCAAAGUCGGCUCGCACCAAUUAAGAGGUGU---GCACGGGCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....((((((.....((.......))....((((((((((((........)))).---...))))))))))))))......))))).. ( -31.30) >consensus AUGAAUUUUCGGCUAACUGUUGAUGCUGCGGUUAUUUUGCAGCCUCGGCUCGCAUCAAUUAAGAGGUGUGCUGCACGGGCCGAGUCAAUUAAGCUGCCGGCA ........(((((.....(((((.(((((((.....)))))))(((((((((((((........)))).......))))))))))))))......))))).. (-33.27 = -33.61 + 0.34)

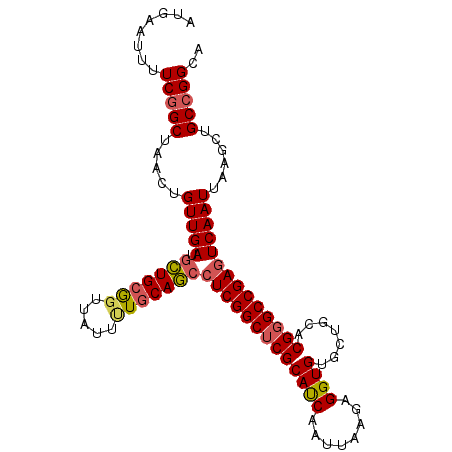
| Location | 11,427,986 – 11,428,088 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -28.55 |
| Energy contribution | -29.17 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11427986 102 - 22407834 UGCCGGCAGCUUAAUUGACUCGGCCCGUGCAGCACACCUCUUAAUUGAUGCGAGCCGAGGCUGCAAAAUAACCGCAGCAUCAACAGUUAGCCGAAAAUUCAU ...((((((((...((((((((((...((((.((...........)).)))).))))))(((((.........))))).)))).)))).))))......... ( -33.40) >DroSec_CAF1 145520 102 - 1 UGCCUGCAGCUUAAUUGACUCGGCCCGUGCAGCGCACCUCUUAAUUGAUGCGAGCCGAGGCUGCAAAAUAACCGCAGCAUCAACAGUUAGCCGAAAAUGCAU (((...(.(((.(((((.((((((.(((((...)))).((......))...).))))))(((((.........))))).....)))))))).).....))). ( -30.60) >DroSim_CAF1 153053 102 - 1 UGCCGGCAGCUUAAUUGACUCGGCCCGUGCAGCACACCUCUUAAUUGAUGCGAGCCGAGGCUGCAAAAUAACCGCAGCAUCAACAGUUAGCCGAAAAUGCAU (((((((((((...((((((((((...((((.((...........)).)))).))))))(((((.........))))).)))).)))).)))).....))). ( -33.60) >DroEre_CAF1 153138 100 - 1 UGCCGGCAGCUUAAUUGACUCGGC-CGUGCAGCACACCUCUUAAUUGAUGCGAGCCGAGGCUGCA-AAUAACCGCAGCAUCAACAGUUAGCCGAAAAUUCAU ...((((((((...((((((((((-..((((.((...........)).)))).))))))(((((.-.......))))).)))).)))).))))......... ( -34.10) >DroYak_CAF1 150961 102 - 1 UGCCGGCAGCUUAAUUGACUCGGCCCGUGCAACACACCUCUUAAUUGAUGCGAGCCGAGGCUGCAGAAUAACUGCAGCAUCAACAGUUAGCCGAAAAUUCAU ...((((((((...((((((((((.(((((((............))).)))).))))))(((((((.....))))))).)))).)))).))))......... ( -38.20) >DroAna_CAF1 130996 99 - 1 UGCCGGCAGCUUAAUUGACUCGGCCCGUGC---ACACCUCUUAAUUGGUGCGAGCCGACUUUGGAAAAUAACCGCAACAUCAACAGUUAGCCGAAAAUUCAU ...((((((((...((((.(((((.((...---.((((........)))))).)))))....((.......))......)))).)))).))))......... ( -25.30) >consensus UGCCGGCAGCUUAAUUGACUCGGCCCGUGCAGCACACCUCUUAAUUGAUGCGAGCCGAGGCUGCAAAAUAACCGCAGCAUCAACAGUUAGCCGAAAAUUCAU ...((((((((...((((((((((.(((((((............))).)))).))))))(((((.........))))).)))).)))).))))......... (-28.55 = -29.17 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:42 2006