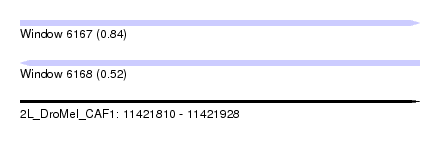
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,421,810 – 11,421,928 |
| Length | 118 |
| Max. P | 0.839432 |

| Location | 11,421,810 – 11,421,928 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -24.97 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
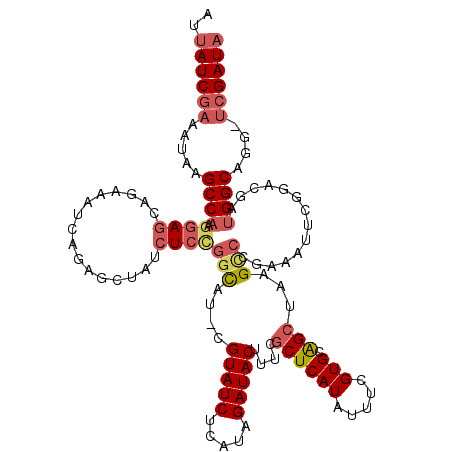
>2L_DroMel_CAF1 11421810 118 + 22407834 UAUCGA-CCUGCCAUCGUCCGAAUUUUGGGCUUAGCUGCACGAAAUAUGAGCAAAAGUAUCUAUGAGAUACG-AUGCCGGAGAUAGCUCUGAUUUCUGCUCCUUGGCUUAUUUCGAUAAU ((((((-...((((..((((((...))))))..((..(((.(((((....(((...(((((.....))))).-.)))(((((....)))))))))))))..)))))).....)))))).. ( -35.90) >DroPse_CAF1 169302 118 + 1 UAUCGAGCCAGCCAUUGUGCCAGGCCCCAGCUUAGCCGCACAAAAUAUGAGCGAAAGUAUCUAUGAGAUACGGAUGGGGGAGUGGG--AGUGGGGGAGAUCCCUGGCUUAUGUCGAUAAU ((((((((((.(((((...(((..((((.(((((.............)))))....(((((.....))))).....))))..))).--)))))(((....))))))).....)))))).. ( -41.22) >DroSec_CAF1 139411 118 + 1 UAUCGA-CCUGCCAUCGUCCGAAUUUCGGACUUAGCUGCACGAAAUAUGAGUAAAAGUAUCUAUGAGAUACG-AUGCCGGAGAUAGCUCUGAUUUCUGCUCCGUGGCUUAUUUCGAUAAU ((((((-...(((((.(((((.....)))))...(..(((.(((((....(((...(((((.....))))).-.)))(((((....)))))))))))))..)))))).....)))))).. ( -37.90) >DroEre_CAF1 147021 118 + 1 UAUCGG-CCUGCCAUCGUUUGGAAUUCGGGCUUAGCUACACGAAAUAUGAGCGAAAGUAUCUAUGAGAUACG-AUGCCAGAGAUAGCUCUGAUUUCUGCUCCUUGGCUUAUGUCGAUAAU ((((((-(..((((((((.(((.............))).)))).....(((((((((((((.....))))).-....(((((....))))).)))).))))..))))....))))))).. ( -35.52) >DroYak_CAF1 144119 118 + 1 UAUCGA-CCUGCCAUCGUCCGAAAUUCGGGCUUAGCUACACGAAAUAUGAGCGAAAGUAUCUAUGAGAUACG-AUGCCAGAGAUAGCUCUGAUUUCUGCUCCUUGGCUUACUUCGAUUAU .(((((-...((((..(((((.....))))).................(((((((((((((.....))))).-....(((((....))))).)))).))))..)))).....)))))... ( -35.30) >DroAna_CAF1 124817 117 + 1 UAUCGA-CCUGCCGCUGUCUGAGGGCC-GGCUUAGCUGCACGAAAUAUGAGCGAAAGUAUCUAUGAGAUACG-AUACCCGAGAUAGGCCGUUCUCGGGCUGCCUGGCUUAUAUUGAUAAU ((((((-...(((((.((((((((((.-(((((((((.((.......)))))....(((((.....))))).-..........)))))))))))))))).))..))).....)))))).. ( -40.50) >consensus UAUCGA_CCUGCCAUCGUCCGAAAUUCGGGCUUAGCUGCACGAAAUAUGAGCGAAAGUAUCUAUGAGAUACG_AUGCCGGAGAUAGCUCUGAUUUCUGCUCCUUGGCUUAUUUCGAUAAU ((((((....((((..(((((.....))))).................((((....(((((.....)))))......(((((....)))))......))))..)))).....)))))).. (-24.97 = -25.75 + 0.78)

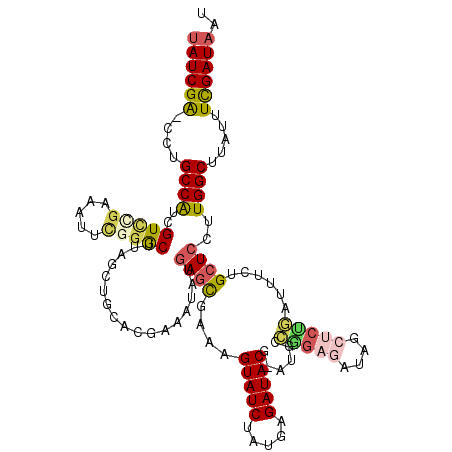
| Location | 11,421,810 – 11,421,928 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -18.09 |
| Energy contribution | -19.21 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11421810 118 - 22407834 AUUAUCGAAAUAAGCCAAGGAGCAGAAAUCAGAGCUAUCUCCGGCAU-CGUAUCUCAUAGAUACUUUUGCUCAUAUUUCGUGCAGCUAAGCCCAAAAUUCGGACGAUGGCAGG-UCGAUA ..((((((.....((((..((((((((....(((((......))).)-)(((((.....))))))))))))).....((((((......))((.......))))))))))...-)))))) ( -33.00) >DroPse_CAF1 169302 118 - 1 AUUAUCGACAUAAGCCAGGGAUCUCCCCCACU--CCCACUCCCCCAUCCGUAUCUCAUAGAUACUUUCGCUCAUAUUUUGUGCGGCUAAGCUGGGGCCUGGCACAAUGGCUGGCUCGAUA ..(((((((...((((((((......)))...--.(((...(((((...(((((.....)))))..((((.((.....)).))))......)))))..))).....))))).).)))))) ( -38.70) >DroSec_CAF1 139411 118 - 1 AUUAUCGAAAUAAGCCACGGAGCAGAAAUCAGAGCUAUCUCCGGCAU-CGUAUCUCAUAGAUACUUUUACUCAUAUUUCGUGCAGCUAAGUCCGAAAUUCGGACGAUGGCAGG-UCGAUA ..((((((.....(((((...((((((((..(((((......)))..-.(((((.....)))))......))..))))).)))......(((((.....)))))).))))...-)))))) ( -33.50) >DroEre_CAF1 147021 118 - 1 AUUAUCGACAUAAGCCAAGGAGCAGAAAUCAGAGCUAUCUCUGGCAU-CGUAUCUCAUAGAUACUUUCGCUCAUAUUUCGUGUAGCUAAGCCCGAAUUCCAAACGAUGGCAGG-CCGAUA ..(((((.(....((((..((((.((..((((((....))))))..)-)(((((.....)))))....))))....((((.((......)).))))..........))))..)-.))))) ( -31.00) >DroYak_CAF1 144119 118 - 1 AUAAUCGAAGUAAGCCAAGGAGCAGAAAUCAGAGCUAUCUCUGGCAU-CGUAUCUCAUAGAUACUUUCGCUCAUAUUUCGUGUAGCUAAGCCCGAAUUUCGGACGAUGGCAGG-UCGAUA ...(((((.....((((..((((.((..((((((....))))))..)-)(((((.....)))))....)))).....((((...((...))(((.....)))))))))))...-))))). ( -34.80) >DroAna_CAF1 124817 117 - 1 AUUAUCAAUAUAAGCCAGGCAGCCCGAGAACGGCCUAUCUCGGGUAU-CGUAUCUCAUAGAUACUUUCGCUCAUAUUUCGUGCAGCUAAGCC-GGCCCUCAGACAGCGGCAGG-UCGAUA ..((((.......(((.(((.((((((((........))))))))..-.(((((.....)))))....((((((.....))).)))...)))-))).....(((........)-)))))) ( -36.10) >consensus AUUAUCGAAAUAAGCCAAGGAGCAGAAAUCAGAGCUAUCUCCGGCAU_CGUAUCUCAUAGAUACUUUCGCUCAUAUUUCGUGCAGCUAAGCCCGAAAUUCGGACGAUGGCAGG_UCGAUA ..((((((.....((((.((((................))))(((....(((((.....)))))....((((((.....))).)))...)))..............))))....)))))) (-18.09 = -19.21 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:35 2006