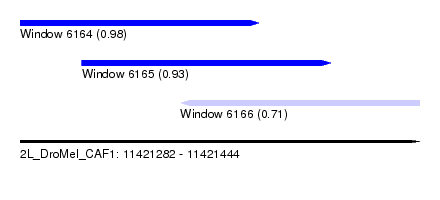
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,421,282 – 11,421,444 |
| Length | 162 |
| Max. P | 0.975830 |

| Location | 11,421,282 – 11,421,379 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11421282 97 + 22407834 --------CAUUC--ACAAUUCCAUU---CUCAGCUG--CUGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUC--UGAUUUGCAACA-GCAUAAGCC----- --------.....--...........---....((((--.((((((((......(((((.....(((((......)))))...)))))...--.)))))))).))-)).......----- ( -23.70) >DroPse_CAF1 168783 114 + 1 CACUCCAACACUCGAGCAACUCCAUU---GAUGGCUG--UUGAAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUCUGAUUUGCAACA-ACAAAAGCCCGAAA ...........(((.((....(((..---..))).((--(((.(((((......(((((.....(((((......)))))...)))))......))))).)))))-......)).))).. ( -21.30) >DroEre_CAF1 146509 100 + 1 --------CAUUC--AGAAAUCCAUUCAACUCAGCUG--CUGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCGCGC--UGAUUUGCAACA-GCAUAAGCC----- --------.....--..................((((--.((((((((......(((((.....(((((......)))))...)))))...--.)))))))).))-)).......----- ( -23.00) >DroYak_CAF1 143591 97 + 1 --------CGUUC--ACAGCUCCGUU---CUCAGCUG--CUGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUC--UGAUUUGCAAGA-GCAUAAGCC----- --------.((((--.(((((.....---...)))))--.((((((((......(((((.....(((((......)))))...)))))...--.)))))))).))-)).......----- ( -25.60) >DroAna_CAF1 124268 100 + 1 --------CAUUU--ACAACUCCAUU---CUCAGCUGUGUUGAAAAUCUCUUUAUGACACAUUUUGAUAUUUAUUUAUUAUCUUGUCACUC--UGAUUUAAAAAACACCCAAGCC----- --------.....--...........---....((((((((..(((((......(((((.....(((((......)))))...)))))...--.)))))....)))))...))).----- ( -13.90) >DroPer_CAF1 168108 114 + 1 CACUCCAACACUCGAGCAACUCCAUU---GAUGGCUG--UUGAAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUUUGAUUGGCAACA-ACAAAAGCCCGAAA ...........(((.((....(((..---..))).((--(((..((((......(((((.....(((((......)))))...)))))......))))..)))))-......)).))).. ( -21.80) >consensus ________CAUUC__ACAACUCCAUU___CUCAGCUG__CUGAAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUC__UGAUUUGCAACA_ACAUAAGCC_____ .................................(((....((.(((((......(((((.....(((((......)))))...)))))......))))).)).........)))...... ( -9.32 = -9.52 + 0.20)

| Location | 11,421,307 – 11,421,408 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -14.49 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11421307 101 + 22407834 UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUGCAACA-GCAUAAGCCACCCUGUUCUUUUUUCCCAUUUUUUUGUG .(((((((......(((((.....(((((......)))))...)))))....)))))))((((-(...........)))))..................... ( -16.20) >DroSec_CAF1 138909 100 + 1 UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUGGAACA-GCAUAAGCCACCCCGUUUUUU-UUUCCAUUUUUUUGUG .(((((........(((((.....(((((......)))))...)))))....((..(((((..-....((((......))))...-.)))))..))))))). ( -13.20) >DroSim_CAF1 145699 98 + 1 UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUGCAACA-GCAUAAGCCACCCCGUUUU---UUUCCAUUUUUUUGUG ((((((((......(((((.....(((((......)))))...)))))....))))))))...-((....))...........---................ ( -15.90) >DroEre_CAF1 146537 96 + 1 UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCGCGCUGAUUUGCAACA-GCAUAAGCCACCCUGUUU----UUCCCAUUUUUUAG-A .(((((((......(((((.....(((((......)))))...)))))....)))))))((((-(...........))))).----..............-. ( -15.50) >DroYak_CAF1 143616 96 + 1 UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUGCAAGA-GCAUAAGCCACCCCGAUU----UUUCCAUUUUUUAG-A ((((((((......(((((.....(((((......)))))...)))))....)))))))).(.-((....))).........----..............-. ( -16.30) >DroAna_CAF1 124295 100 + 1 UGAAAAUCUCUUUAUGACACAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUAAAAAACACCCAAGCCACCCCCAUUUUC-AGGACAUUUCAUUC-A (((((.........(((((.....(((((......)))))...))))).(((((.............................))-)))...)))))...-. ( -9.85) >consensus UGCAAAUCUCCUUAUGACAUAUUUUGAUAUUUAUUUAUUAUCUUGUCACUCUGAUUUGCAACA_GCAUAAGCCACCCCGUUUU___UUUCCAUUUUUUUG_A ((((((((......(((((.....(((((......)))))...)))))....)))))))).......................................... (-12.82 = -13.18 + 0.36)

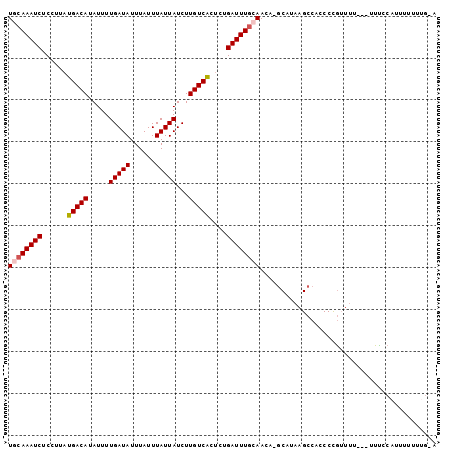
| Location | 11,421,347 – 11,421,444 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -16.63 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
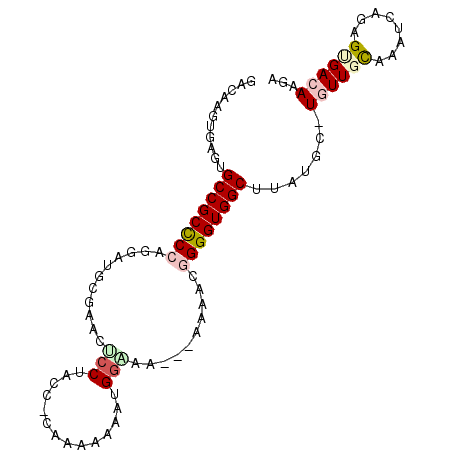
>2L_DroMel_CAF1 11421347 97 - 22407834 GACAAGUGAGUGCCGCCCCAGGAUGCGAAGCCCUACCACAAAAAAAUGGGAAAAAAGAACAGGGUGGCUUAUGC-UGUUGCAAAUCAGAGUGACAAGA ....((((...(((((((.(((.........))).(((........)))............)))))))...)))-)((..(........)..)).... ( -22.80) >DroSec_CAF1 138949 96 - 1 GACAAGUGAGUGCCGCCCCAGGAUGCUAACUCCUACCACAAAAAAAUGGAAA-AAAAAACGGGGUGGCUUAUGC-UGUUCCAAAUCAGAGUGACAAGA ...........((((((((((((.......)))).(((........)))...-.......))))))))((((.(-((........))).))))..... ( -26.10) >DroSim_CAF1 145739 94 - 1 GACAAGUGAGUGCCGCCCCAGGAUGCUAACUCCUACCACAAAAAAAUGGAAA---AAAACGGGGUGGCUUAUGC-UGUUGCAAAUCAGAGUGACAAGA ....((((...((((((((((((.......)))).(((........)))...---.....))))))))...)))-)((..(........)..)).... ( -28.60) >DroEre_CAF1 146577 92 - 1 GACAAGUGAAUGCCGCCCCAGGAUGCGAAGUCCUAGU-CUAAAAAAUGGGAA----AAACAGGGUGGCUUAUGC-UGUUGCAAAUCAGCGCGACAAGA ....((((...(((((((.(((((.....)))))..(-((........))).----.....)))))))...)))-)(((((........))))).... ( -27.20) >DroYak_CAF1 143656 92 - 1 GACAAGUGAGUGCCGCCCCAGGAUGCGAAUCCCUGCU-CUAAAAAAUGGAAA----AAUCGGGGUGGCUUAUGC-UCUUGCAAAUCAGAGUGACAAGA ..((..(((..((((((((.((((....))))....(-(((.....))))..----....))))))))...(((-....)))..)))...))...... ( -27.40) >DroAna_CAF1 124335 86 - 1 GACAAGUGAGCGCCGCUCCU----------UCCUUCU-GAAUGAAAUGUCCU-GAAAAUGGGGGUGGCUUGGGUGUUUUUUAAAUCAGAGUGACAAGA ...........(((((((((----------...(((.-((........))..-)))...))))))))).....((((.(((......))).))))... ( -18.90) >consensus GACAAGUGAGUGCCGCCCCAGGAUGCGAACUCCUACC_CAAAAAAAUGGAAA___AAAACGGGGUGGCUUAUGC_UGUUGCAAAUCAGAGUGACAAGA ...........((((((((...........(((..............)))..........)))))))).......((((((........))))))... (-16.63 = -17.57 + 0.95)

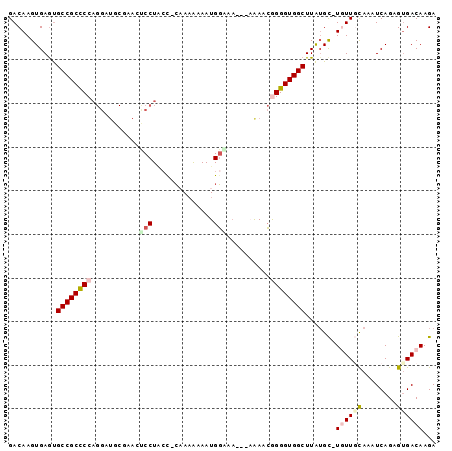
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:33 2006