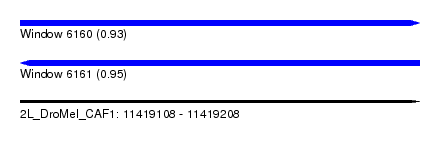
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,419,108 – 11,419,208 |
| Length | 100 |
| Max. P | 0.947979 |

| Location | 11,419,108 – 11,419,208 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -14.62 |
| Energy contribution | -16.54 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
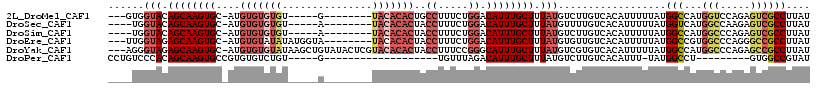
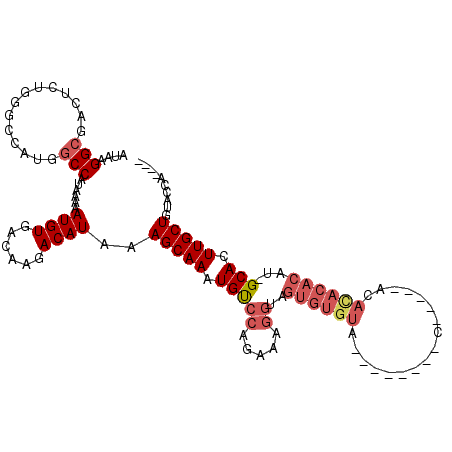
>2L_DroMel_CAF1 11419108 100 + 22407834 ---GUGGUACAGCAAGUGC-AUGUGUGUGU-----G--------UACACACUGCCUUUCUGGACAUUUGCUUUAUGUCUUGUCACAUUUUUAUGGCCAUGGUCCAGAGUCGCCUUAU ---..(((((...(((.((-(.((((((..-----.--------.))))))))))))(((((((....(((..((((......))))......)))....))))))))).))).... ( -30.40) >DroSec_CAF1 136696 99 + 1 ----UGGUACAGCAAGUGC-AUGUGUGUGU-----A--------UACACACUACCUUUCUGGACAUUUGCUUUAUGUUUUGUCACAUUUUUAUGGUCAUGGCCAAGAGUCGCCUUAU ----.(((((((((((((.-..((((((..-----.--------.))))))..((.....)).)))))))).....................((((....))))...)).))).... ( -25.00) >DroSim_CAF1 143494 99 + 1 ----UGGUACAGCAAGUGC-AUGUGUGUGU-----A--------UACACACUACCUUUCUGGACAUUUGCUUUAUGUCUUGUCACAUUUUUAUGGCCAUGGCCCAGAGUCGCCUUAU ----.((((..((....))-..((((((..-----.--------.)))))))))).(((((((((...((.....))..))))..........(((....))))))))......... ( -25.20) >DroEre_CAF1 144435 105 + 1 ---UUGGUAGAGCAAGUGC-AUGUGUAUAUAUGGUA--------UACACACUACCUUUCUGGACAUUUGCUUUAUGUGUUGUCACAUUUUUAUGGCCGUGGCCCAGGGCCGCCUUAU ---..(((((((((((((.-.(((((((((...)))--------))))))...((.....)).))))))))))(((((....))))).......)))((((((...))))))..... ( -36.30) >DroYak_CAF1 141296 113 + 1 ---AGGGUAGAGCAAGUGC-AUGUGUGUAUAAGCUGUAUACUCGUACACACUACCUUUCCGGGCAUUUGCUUUAUGUCGUGUCACAUUUUUAUGGCCAUGGCCCAGAGCCGCCUUAU ---.(((((((((((((((-..(((((((..((.......))..)))))))..((.....)))))))))))))))((((((.(.(((....)))).))))))))............. ( -39.30) >DroPer_CAF1 165705 83 + 1 CCUGUCCCACAGCAAGUGCCGUGUGUCUGU-----G-------------------UGUUUAGACAUUUGCUUUAUGUCUUGUCACAUUU-UAUGGCCU---------GUGGCCGUAU ......((((((.....((((((.....((-----(-------------------((...((((((.......))))))...)))))..-))))))))---------))))...... ( -26.60) >consensus ____UGGUACAGCAAGUGC_AUGUGUGUGU_____A________UACACACUACCUUUCUGGACAUUUGCUUUAUGUCUUGUCACAUUUUUAUGGCCAUGGCCCAGAGUCGCCUUAU ......(((.((((((((....(((((((...............)))))))..((.....)).)))))))).)))..................(((...(((.....)))))).... (-14.62 = -16.54 + 1.92)
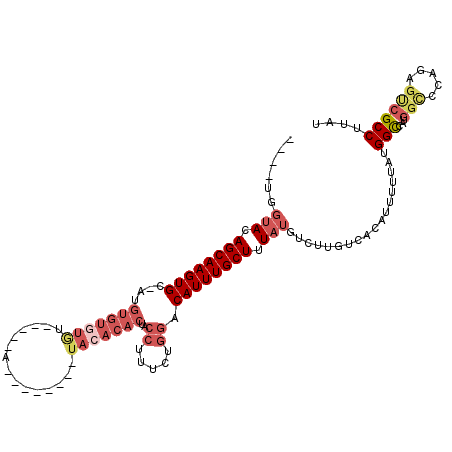
| Location | 11,419,108 – 11,419,208 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -12.45 |
| Energy contribution | -13.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11419108 100 - 22407834 AUAAGGCGACUCUGGACCAUGGCCAUAAAAAUGUGACAAGACAUAAAGCAAAUGUCCAGAAAGGCAGUGUGUA--------C-----ACACACACAU-GCACUUGCUGUACCAC--- ....(((...(((((((....((.......((((......))))...))....)))))))((((((((((((.--------.-----..)))))).)-)).)))))).......--- ( -28.00) >DroSec_CAF1 136696 99 - 1 AUAAGGCGACUCUUGGCCAUGACCAUAAAAAUGUGACAAAACAUAAAGCAAAUGUCCAGAAAGGUAGUGUGUA--------U-----ACACACACAU-GCACUUGCUGUACCA---- ....(((........)))............((((......)))).........((.(((.((((((((((((.--------.-----..)))))).)-)).))).))).))..---- ( -21.30) >DroSim_CAF1 143494 99 - 1 AUAAGGCGACUCUGGGCCAUGGCCAUAAAAAUGUGACAAGACAUAAAGCAAAUGUCCAGAAAGGUAGUGUGUA--------U-----ACACACACAU-GCACUUGCUGUACCA---- ....(((...(((((((....)))...............(((((.......)))))))))((((((((((((.--------.-----..)))))).)-)).))))))......---- ( -26.50) >DroEre_CAF1 144435 105 - 1 AUAAGGCGGCCCUGGGCCACGGCCAUAAAAAUGUGACAACACAUAAAGCAAAUGUCCAGAAAGGUAGUGUGUA--------UACCAUAUAUACACAU-GCACUUGCUCUACCAA--- ....(((((((...))))...)))......(((((....)))))..(((((.(((((.....))..(((((((--------((.....)))))))))-))).))))).......--- ( -28.30) >DroYak_CAF1 141296 113 - 1 AUAAGGCGGCUCUGGGCCAUGGCCAUAAAAAUGUGACACGACAUAAAGCAAAUGCCCGGAAAGGUAGUGUGUACGAGUAUACAGCUUAUACACACAU-GCACUUGCUCUACCCU--- ...(((.((((((((((....((.......((((......))))...))....)))))))(((((((((((((..(((.....)))..))))))).)-)).))))))....)))--- ( -34.00) >DroPer_CAF1 165705 83 - 1 AUACGGCCAC---------AGGCCAUA-AAAUGUGACAAGACAUAAAGCAAAUGUCUAAACA-------------------C-----ACAGACACACGGCACUUGCUGUGGGACAGG ....((((..---------.))))...-...((((...((((((.......))))))...))-------------------)-----)......((((((....))))))....... ( -24.00) >consensus AUAAGGCGACUCUGGGCCAUGGCCAUAAAAAUGUGACAAGACAUAAAGCAAAUGUCCAGAAAGGUAGUGUGUA________C_____ACACACACAU_GCACUUGCUGUACCA____ ....(((..............)))......((((......))))..(((((.(((((.....))..((((((.................))))))...))).))))).......... (-12.45 = -13.70 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:28 2006