
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
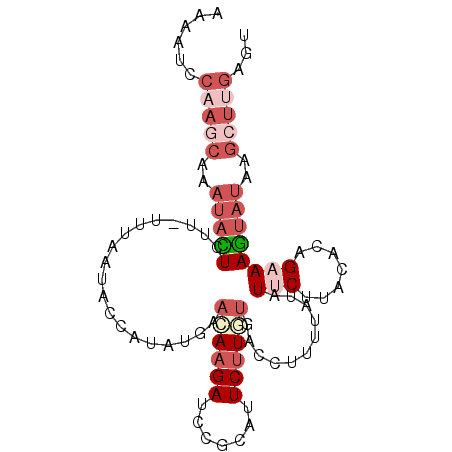
| Location | 11,417,032 – 11,417,126 |
| Length | 94 |
| Max. P | 0.749866 |

| Location | 11,417,032 – 11,417,126 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -16.37 |
| Consensus MFE | -2.58 |
| Energy contribution | -5.22 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
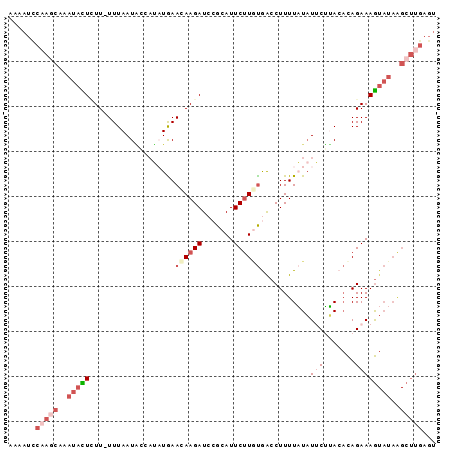
>2L_DroMel_CAF1 11417032 94 + 22407834 AAAAUCCAAGCAAAUACUGUU-UUUAAUAACAUAUGAAAAAGAUCCGCAUUCUUGUGACCUUUUAUAUUCUUACACAGAAAGUAUAAGCUUGAGU ......(((((..(((((...-...............(((((...((((....))))..)))))...((((.....)))))))))..)))))... ( -15.50) >DroSec_CAF1 134539 94 + 1 AAAAUUCAAGCAAAUACUUUU-UUUAAUGUCAUAUAAAUAAGAUCCGCAUUCUUAUGACCUUUUAUAUGCUUACACAGAAAGUAUAAGCUUGAGU ...((((((((..((((((((-(.....(.(((((((((((((.......))))))......))))))))......)))))))))..)))))))) ( -23.00) >DroSim_CAF1 141340 94 + 1 AAAAUCCAAGCAAAUACUGUU-UUUAAUGCCAUAUAAAUAAGAUCCGCAUUCUUCUGACCUUUUAUAUGCUUACACAGAAAGUAUAAGCUUGAGU ......(((((..(((((...-........((((((((..(((..........))).....))))))))...........)))))..)))))... ( -16.15) >DroEre_CAF1 142155 81 + 1 GAAAUCGUAGCAAAUAUUCUU-UUUAAUACCAAUUGAACAAGAUCCGCAUUCUUGCAAACUUUUUUCUUCGGACUUAGAAAA------------- .................((((-((((((....)))))).))))...(((....)))......((((((........))))))------------- ( -8.00) >DroYak_CAF1 139048 95 + 1 AAAAUCCUAUCAAAUAUUCUUGUUUAAUUCCCAUUGAACAAGAUCCGCAUUCCUGUAGCCUUUUCUAUUCGGACACAGAAAGUAUAGGCUUGAGU .....((((((......(((((((((((....))))))))))).........((((..((..........))..))))...).)))))....... ( -19.20) >consensus AAAAUCCAAGCAAAUACUCUU_UUUAAUACCAUAUGAACAAGAUCCGCAUUCUUGUGACCUUUUAUAUUCUUACACAGAAAGUAUAAGCUUGAGU ......(((((..(((((...................((((((.......))))))...........(((.......))))))))..)))))... ( -2.58 = -5.22 + 2.64)

| Location | 11,417,032 – 11,417,126 |
|---|---|
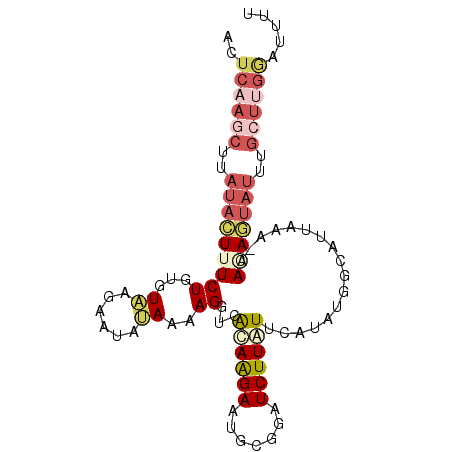
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -4.20 |
| Energy contribution | -6.44 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.22 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
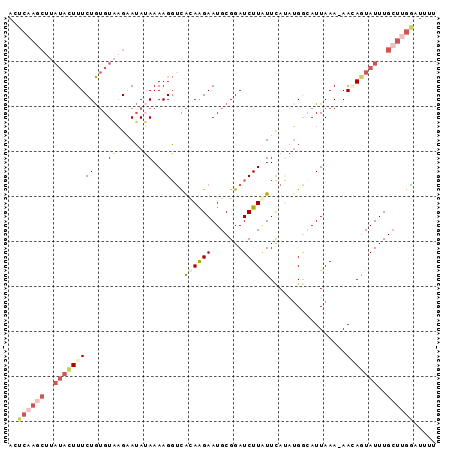
>2L_DroMel_CAF1 11417032 94 - 22407834 ACUCAAGCUUAUACUUUCUGUGUAAGAAUAUAAAAGGUCACAAGAAUGCGGAUCUUUUUCAUAUGUUAUUAAA-AACAGUAUUUGCUUGGAUUUU ..((((((..(((((......((((..((((((((((((.(........)))))))))..)))).))))....-...)))))..))))))..... ( -19.32) >DroSec_CAF1 134539 94 - 1 ACUCAAGCUUAUACUUUCUGUGUAAGCAUAUAAAAGGUCAUAAGAAUGCGGAUCUUAUUUAUAUGACAUUAAA-AAAAGUAUUUGCUUGAAUUUU ..((((((..(((((((..((((...(((((((((((((...........)))))..))))))))))))....-.)))))))..))))))..... ( -24.20) >DroSim_CAF1 141340 94 - 1 ACUCAAGCUUAUACUUUCUGUGUAAGCAUAUAAAAGGUCAGAAGAAUGCGGAUCUUAUUUAUAUGGCAUUAAA-AACAGUAUUUGCUUGGAUUUU ..((((((..(((((((..((((...(((((((((((((.(.......).)))))..))))))))))))..))-...)))))..))))))..... ( -21.60) >DroEre_CAF1 142155 81 - 1 -------------UUUUCUAAGUCCGAAGAAAAAAGUUUGCAAGAAUGCGGAUCUUGUUCAAUUGGUAUUAAA-AAGAAUAUUUGCUACGAUUUC -------------......(((((....(((.((..((((((....))))))..)).)))...(((((.....-.........))))).))))). ( -10.24) >DroYak_CAF1 139048 95 - 1 ACUCAAGCCUAUACUUUCUGUGUCCGAAUAGAAAAGGCUACAGGAAUGCGGAUCUUGUUCAAUGGGAAUUAAACAAGAAUAUUUGAUAGGAUUUU .......(((((..((((((((.((..........)).))))))))......(((((((.(((....))).))))))).......)))))..... ( -21.60) >consensus ACUCAAGCUUAUACUUUCUGUGUAAGAAUAUAAAAGGUCACAAGAAUGCGGAUCUUAUUCAUAUGGCAUUAAA_AACAGUAUUUGCUUGGAUUUU ..((((((..(((((((((...((......))..))...((((((.......)))))).................)))))))..))))))..... ( -4.20 = -6.44 + 2.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:25 2006