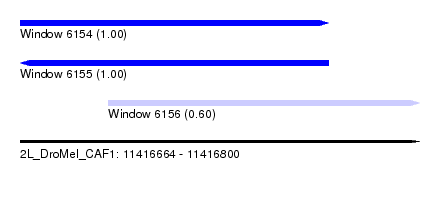
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,416,664 – 11,416,800 |
| Length | 136 |
| Max. P | 0.999578 |

| Location | 11,416,664 – 11,416,769 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -21.93 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11416664 105 + 22407834 UUUACUUUUUUAU-----GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU--AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC .............-----........((((....)))).((((..(.....(((((((.........))))))).--)..)))).....((((((((....))))))))... ( -22.70) >DroSec_CAF1 134170 105 + 1 UUUACUUUUUUAU-----GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUAACAAU--AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC ...((((...(((-----.(((....((((....))))....((((.........))))...........)))))--)...))))....((((((((....))))))))... ( -21.10) >DroSim_CAF1 140971 105 + 1 UUUACUUUUUUAU-----GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU--AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC .............-----........((((....)))).((((..(.....(((((((.........))))))).--)..)))).....((((((((....))))))))... ( -22.70) >DroEre_CAF1 141799 105 + 1 CUUACUUUUUUAU-----GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUGUACAAU--AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC .............-----........((((....)))).((((..(.....((((..((((.....)))))))).--)..)))).....((((((((....))))))))... ( -22.70) >DroYak_CAF1 138681 105 + 1 UUUACUUUUUUAU-----GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU--AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC .............-----........((((....)))).((((..(.....(((((((.........))))))).--)..)))).....((((((((....))))))))... ( -22.70) >DroAna_CAF1 120127 112 + 1 UUUACUUUUUUAUGGUGUGUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAUAUAUUUGAGACAGAGCUGCAGAUGCAGCUGCAGCAUC ...........(((.(((.((((((.((((....)))).............(((((((.........))))))).......)))))).((((((....))))))))).))). ( -27.20) >consensus UUUACUUUUUUAU_____GUGUCUUCGCCUCGUUGGGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU__AUUCGAGUCUGAGCUGCAGAUGCAUCUGCAGCAUC ........................((((((....)))).((((........(((((((.........)))))))......))))...))(((((((......)))))))... (-21.93 = -22.12 + 0.20)

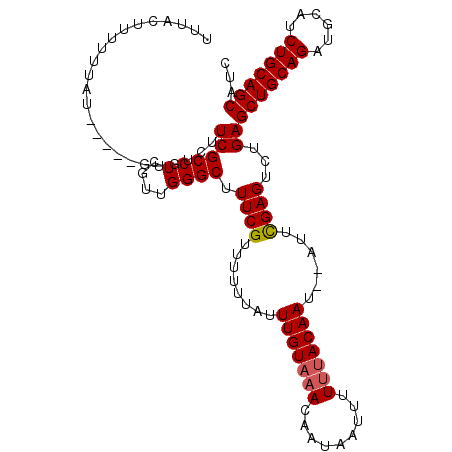
| Location | 11,416,664 – 11,416,769 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11416664 105 - 22407834 GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU--AUUGUAAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC-----AUAAAAAAGUAAA ((.((((((((....))))))))))...((((...--.(((((((.........)))))))........(....).....))))..........-----............. ( -23.70) >DroSec_CAF1 134170 105 - 1 GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU--AUUGUUAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC-----AUAAAAAAGUAAA ((.((((((((....))))))))))..(((....(--((((((........((((((.........))))))..(((......)))...)))).-----)))....)))... ( -22.50) >DroSim_CAF1 140971 105 - 1 GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU--AUUGUAAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC-----AUAAAAAAGUAAA ((.((((((((....))))))))))...((((...--.(((((((.........)))))))........(....).....))))..........-----............. ( -23.70) >DroEre_CAF1 141799 105 - 1 GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU--AUUGUACAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC-----AUAAAAAAGUAAG ((.((((((((....))))))))))...((((...--.((((((((.....))))..))))........(....).....))))..........-----............. ( -22.80) >DroYak_CAF1 138681 105 - 1 GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU--AUUGUAAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC-----AUAAAAAAGUAAA ((.((((((((....))))))))))...((((...--.(((((((.........)))))))........(....).....))))..........-----............. ( -23.70) >DroAna_CAF1 120127 112 - 1 GAUGCUGCAGCUGCAUCUGCAGCUCUGUCUCAAAUAUAUUGUAAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACACACACCAUAAAAAAGUAAA ((.(((((((......)))))))))((((((.......(((((((.........))))))).............(((......)))).)))))................... ( -24.10) >consensus GAUGCUGCAGAUGCAUCUGCAGCUCAGACUCGAAU__AUUGUAAAAAAUUAUUGUUUACAAAUAAAAAACGAAAGCCCAACGAGGCGAAGACAC_____AUAAAAAAGUAAA ((.((((((((....))))))))))...((((......(((((((.........)))))))........(....).....))))............................ (-21.02 = -21.68 + 0.67)

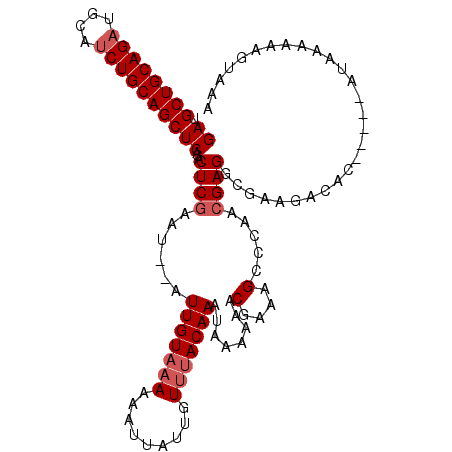
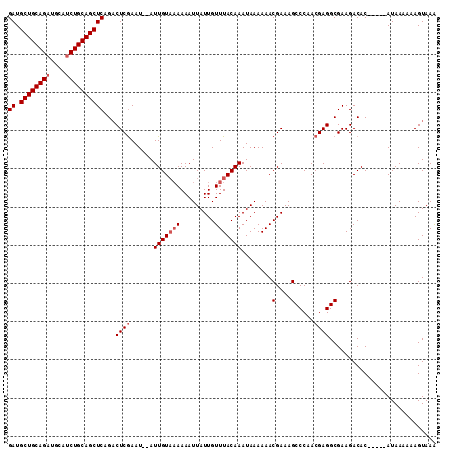
| Location | 11,416,694 – 11,416,800 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -12.47 |
| Energy contribution | -13.97 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11416694 106 + 22407834 GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU--AUUCGAGUCUGAGCUGC---AGAUGCAUCUGCAGCAUCAGCA----CUCCGCCA--UCUAUCUUGUGGAUAC---UUG (((.............(((((((.........))))))).--....(((((((((((((---(((....)))))))).)))).)----))).)))(--(((((...))))))..---... ( -33.60) >DroSec_CAF1 134200 106 + 1 GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUAACAAU--AUUCGAGUCUGAGCUGC---AGAUGCAUCUGCAGCAUCAGCA----CUCCGCCA--UCUAUCUUGUGGAUAC---UUG (((....(((......(((....)))........)))...--....(((((((((((((---(((....)))))))).)))).)----))).)))(--(((((...))))))..---... ( -31.74) >DroEre_CAF1 141829 106 + 1 GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUGUACAAU--AUUCGAGUCUGAGCUGC---AGAUGCAUCUGCAGCAUCAGCA----CUCCGCCA--UCUAUCUUGUGGAUAC---UUG (((.............((((..((((.....)))))))).--....(((((((((((((---(((....)))))))).)))).)----))).)))(--(((((...))))))..---... ( -33.60) >DroYak_CAF1 138711 106 + 1 GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU--AUUCGAGUCUGAGCUGC---AGAUGCAUCUGCAGCAUCAGCA----CUCCGCCA--UCUAUCUUGUGGAUAC---UUG (((.............(((((((.........))))))).--....(((((((((((((---(((....)))))))).)))).)----))).)))(--(((((...))))))..---... ( -33.60) >DroAna_CAF1 120162 115 + 1 GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAUAUAUUUGAGACAGAGCUGC---AGAUGCAGCUGCAGCAUCAGCAUGAGCAUCAGCA--UCUAUCUUGUGGAUGCUACUGC .(((...(((((.((((((((((.........))))))).)))...))))).(((((((---((......))))))).)))))....(((..((((--(((((...)))))))))..))) ( -35.10) >DroPer_CAF1 162228 93 + 1 ----GUUGUUUUUUAUCUGUAAACAAUAAUUUUUUACAAU--AUUUCA-----AGGGUCGAGAGAUGCAUCU-CUCCAUC------------UCCAUCUCCAUCUUGUGGAUAC---UUG ----(((((((.........))))))).............--....((-----(((((.((((((....)))-))).)))------------......(((((...)))))..)---))) ( -17.70) >consensus GGCUUUCGUUUUUUAUUUGUAAACAAUAAUUUUUUACAAU__AUUCGAGUCUGAGCUGC___AGAUGCAUCUGCAGCAUCAGCA____CUCCGCCA__UCUAUCUUGUGGAUAC___UUG (((.((((........(((((((.........)))))))......)))).(((((((((...((......))))))).))))..........)))...(((((...)))))......... (-12.47 = -13.97 + 1.50)

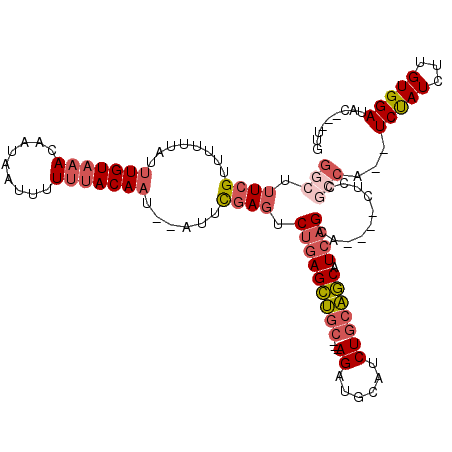
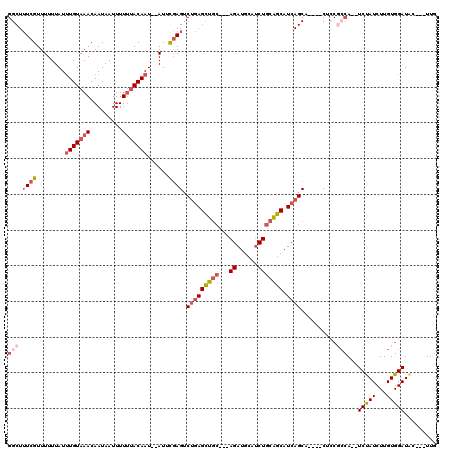
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:24 2006