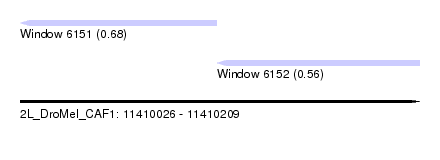
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,410,026 – 11,410,209 |
| Length | 183 |
| Max. P | 0.677768 |

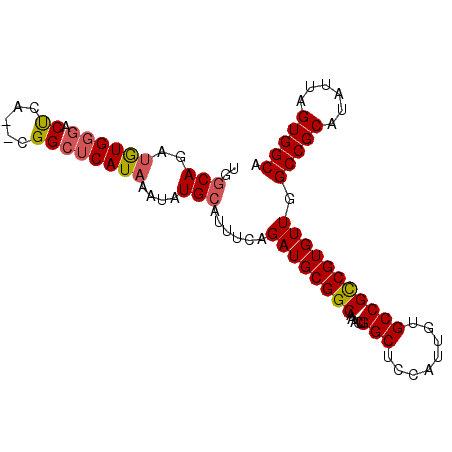
| Location | 11,410,026 – 11,410,116 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11410026 90 - 22407834 AAUAUUGCUGGUCGCCAUAAAAGCCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGUCCACUU-----CGUC-CACUUAAUCAAAGGCCAGUUGCA-- ......(((((((((.......))(((((.((((((((...))))))))..))))).......-----....-............)))))))....-- ( -21.40) >DroPse_CAF1 156484 88 - 1 AAUAUUGCUGGCCACAAUAAAAACCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGCUCGUUCUGCAGCGCAGCUGUUGAUCAAUGCC---------- .........(((............(((((.((((((((...))))))))..)))))...(.((.(((((...))))).)).)...)))---------- ( -20.10) >DroSec_CAF1 127414 90 - 1 AAUAUUGCUGGUCGCCAUAAAAGCCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGUUCACUU-----CGUC-CACUUAAUCAAAGGCCAGUUGCA-- ......(((((((((.......))(((((.((((((((...))))))))..))))).......-----....-............)))))))....-- ( -21.40) >DroSim_CAF1 134191 90 - 1 AAUAUUGCUGGUCGCCAUAAAGGCCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGUUCACUU-----CGUC-CACUUAAUCAAAGGCCAGUUGCA-- ......((((((((((.....)))(((((.((((((((...))))))))..))))).......-----....-............)))))))....-- ( -25.50) >DroEre_CAF1 134924 90 - 1 AAUAUUGCUGGUCGCCAUGAAAGCCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGU-CGCUC-----CGCAGUUGUUAAUCAAAGGCCAGUUGCA-- ......(((((((((...((..(.(((((.((((((((...))))))))..))))).-)..))-----.))..(((.....))).)))))))....-- ( -24.10) >DroYak_CAF1 131886 93 - 1 AAUAUUGCUGGUCGCCAUGAAAGCUGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCCUCCGCUC-----CGCAGUUGUUAAUCAAAGGCCAGUUGCACU ......(((((((....(((.((((((...((((((((...))))))))....((....))..-----.)))))).....)))..)))))))...... ( -22.70) >consensus AAUAUUGCUGGUCGCCAUAAAAGCCGCAUUGUGAAAAUGAAAUUUUCAUUUAUGCGUUCACUC_____CGCA_CACUUAAUCAAAGGCCAGUUGCA__ ......(((((((...........(((((.((((((((...))))))))..)))))...((........))..............)))))))...... (-17.54 = -17.90 + 0.36)

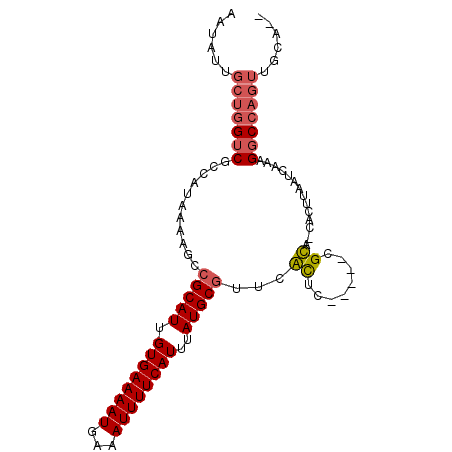
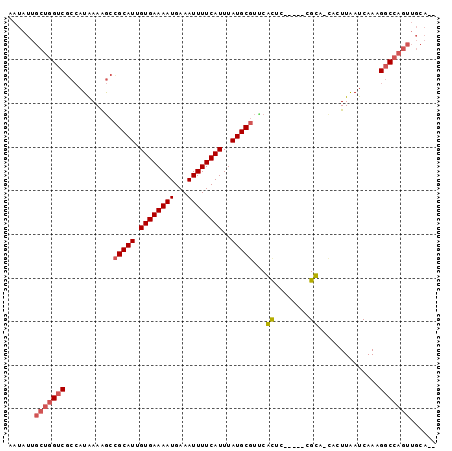
| Location | 11,410,116 – 11,410,209 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -27.75 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11410116 93 - 22407834 UGGCAGAUGUGGGACUCA--CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAAACUGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA ..((.((((((((...((--((((..........(((((...)))))((((...((....))...))))))))))....)).))))))....)). ( -32.60) >DroSec_CAF1 127504 93 - 1 UGGCAGAUGUGGGACUCA--CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAAACUGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA ..((.((((((((...((--((((..........(((((...)))))((((...((....))...))))))))))....)).))))))....)). ( -32.60) >DroSim_CAF1 134281 93 - 1 UGGCAGAUGUGGGACUCA--CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAAACUGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA ..((.((((((((...((--((((..........(((((...)))))((((...((....))...))))))))))....)).))))))....)). ( -32.60) >DroEre_CAF1 135014 93 - 1 UGCCAGAUGUGGGACCCA--CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAGGCCGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA .((((((((((((.((((--((((..........(((((...)))))((....))(((........)))))))))..)))).))))))..)))). ( -37.00) >DroYak_CAF1 131979 93 - 1 UGGCAGAUGUGGGACUCA--CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAAACUGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA ..((.((((((((...((--((((..........(((((...)))))((((...((....))...))))))))))....)).))))))....)). ( -32.60) >DroAna_CAF1 114549 95 - 1 UGGCACUGAUGCCACUCAACGGGCUCAUAAAUAUGCAUUUCAGAUGCGGCUCUUUGGCUCCAUUGUGCCGUCGUGUUGGCCGCAUAUUAGUGGCA (((((....)))))..((((((((..........(((((...)))))(((.(..((....))..).)))))).)))))(((((......))))). ( -32.70) >consensus UGGCAGAUGUGGGACUCA__CGGCUCAUAAAUAUGCAUUUCAGAUGCGGCAAACUGGCUCCAUUGUGCCGCCGUGUUGGCCGCAUAUUAGUGGCA ..(((..((((((.((.....))))))))....)))......((((((((.....(((........))))))))))).(((((......))))). (-27.75 = -28.00 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:20 2006