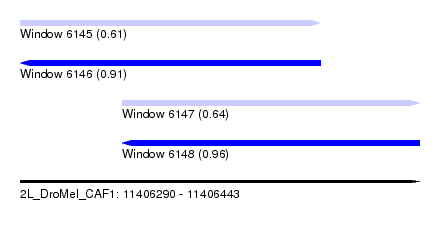
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,406,290 – 11,406,443 |
| Length | 153 |
| Max. P | 0.956315 |

| Location | 11,406,290 – 11,406,405 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11406290 115 + 22407834 UGGUAUGAGGUCCCCGGUGGUAAAAAAAAAACAAAAAAAUUAACUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGCUUGCAAGUCGCAAAGUAAUCGAUUGCUGUUGAAAGGACA .........(((((((.((((.....................))))....))).....((((.(((((((((.(((((((.....)).))))))))))))))))))....)))). ( -26.30) >DroSec_CAF1 123780 103 + 1 UGGUAUGAGGUCCCCGGUGGUAAAA-----------AAAAUAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAGUCGCA-GGUAAUCGAUUACUGUUGAAAGGACA .........(((((((.........-----------..............))).....((((.(((((((((.((.((((.....)))-).)))))))))))))))....)))). ( -23.90) >DroSim_CAF1 130506 108 + 1 UGGUAUGAGGUCCCCGGUGGUAAAA------UAAAAAAAUGAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAUUCGCA-GGUAAUCGAUUACUGUUGAAAGGACA .........(((((((.........------...................))).....((((.(((((((((.((.((((.....)))-).)))))))))))))))....)))). ( -23.69) >DroEre_CAF1 131373 99 + 1 UGGUACGAGGUCCCCGGCGGUAAA------------AAGCUGAGGAAAAACGGGUG---AUGUGUAAUCGAUUUGGUGGCAAGUCGCA-GGUAAUCGAUUGCUGUUGAAAGAACA ..((.(.(.(((((((.((((...------------..))))........)))).)---)).)((((((((((..((((....)))).-...))))))))))........).)). ( -26.40) >consensus UGGUAUGAGGUCCCCGGUGGUAAAA___________AAAUUAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAGUCGCA_GGUAAUCGAUUACUGUUGAAAGGACA .........(((((((..................................)))..........(((((((((.....(((.....))).....)))))))))........)))). (-19.31 = -19.55 + 0.25)

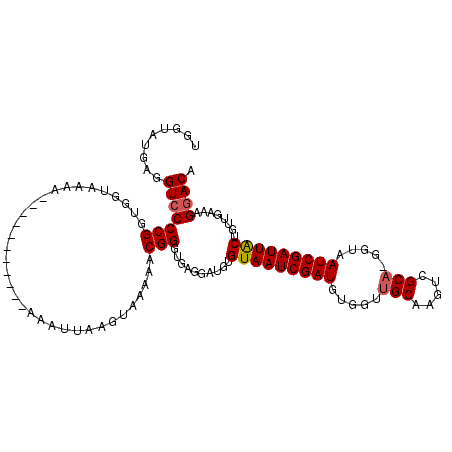
| Location | 11,406,290 – 11,406,405 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11406290 115 - 22407834 UGUCCUUUCAACAGCAAUCGAUUACUUUGCGACUUGCAAGCACAUCGAUUACACAUCCUCACCCGUUUUUAGUUAAUUUUUUUGUUUUUUUUUUACCACCGGGGACCUCAUACCA .((((........(.(((((((...(((((.....)))))...))))))).)..........(((....(((..((............))..)))....)))))))......... ( -16.20) >DroSec_CAF1 123780 103 - 1 UGUCCUUUCAACAGUAAUCGAUUACC-UGCGACUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUAUUUU-----------UUUUACCACCGGGGACCUCAUACCA .((((........(((((((((....-(((.....))).....)))))))))..........(((..............-----------.........)))))))......... ( -18.60) >DroSim_CAF1 130506 108 - 1 UGUCCUUUCAACAGUAAUCGAUUACC-UGCGAAUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUCAUUUUUUUA------UUUUACCACCGGGGACCUCAUACCA .((((........(((((((((....-(((.....))).....)))))))))..........(((...................------.........)))))))......... ( -18.39) >DroEre_CAF1 131373 99 - 1 UGUUCUUUCAACAGCAAUCGAUUACC-UGCGACUUGCCACCAAAUCGAUUACACAU---CACCCGUUUUUCCUCAGCUU------------UUUACCGCCGGGGACCUCGUACCA .............(.((((((((...-.((.....)).....)))))))).)....---....((....(((((.((..------------......)).)))))...))..... ( -16.00) >consensus UGUCCUUUCAACAGCAAUCGAUUACC_UGCGACUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUAAUUU___________UUUUACCACCGGGGACCUCAUACCA .(((.........(((((((((.....(((.....))).....))))))))).........((((..................................)))))))......... (-15.37 = -15.93 + 0.56)

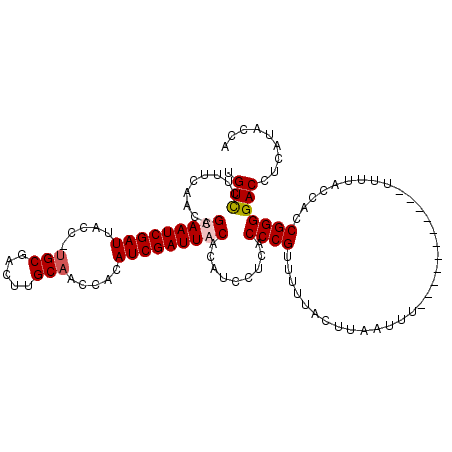
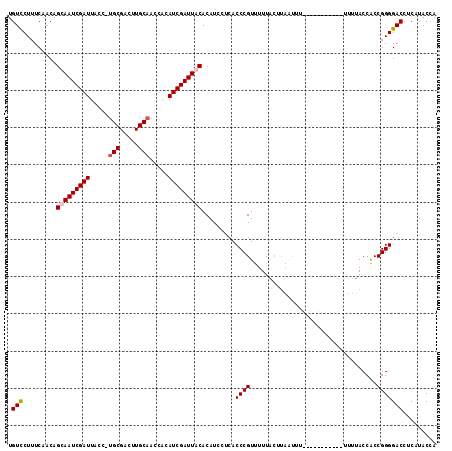
| Location | 11,406,329 – 11,406,443 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11406329 114 + 22407834 UUAACUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGCUUGCAAGUCGCAAAGUAAUCGAUUGCUGUUGAAAGGACACUGCCUCAAGGCGAAAACAGAAACAAUAUUAUUCAACA ............((((((...((((((((((((.(((((((.....)).))))))))))))))((((...((....))(((....)))...))))...)))...)))))).... ( -27.20) >DroSec_CAF1 123808 113 + 1 AUAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAGUCGCA-GGUAAUCGAUUACUGUUGAAAGGACAUUGCCUCAAGGCGAAAACAGAAACAAUAUUAUUCAACA ............((((((...((((((((((((.((.((((.....)))-).)))))))))))((((.....))))(((((....)))))........)))...)))))).... ( -27.30) >DroSim_CAF1 130539 113 + 1 UGAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAUUCGCA-GGUAAUCGAUUACUGUUGAAAGGACAAUGCCUCAAGGCGAAAACAGAAAAAAUAUUAUUCAACA ...........((..(((((....(((((((((.((.((((.....)))-).)))))))))))((((.....))))...)))))...))......................... ( -24.90) >DroEre_CAF1 131400 109 + 1 CUGAGGAAAAACGGGUG---AUGUGUAAUCGAUUUGGUGGCAAGUCGCA-GGUAAUCGAUUGCUGUUGAAAGAACAACGCCUCGAGGCGAAAACAGUAGC-AAAUUAUUCAACA ............(((((---((.(((((((((((..((((....)))).-...))))))))(((((((......)..((((....))))..)))))).))-).))))))).... ( -28.90) >DroYak_CAF1 128013 111 + 1 UUAAAGAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAGUCGCA-GGUGAUCGAUUACUGUUGAAAGGACAAUGCCUCAA-GCGAGAACAGUAAA-AUAUUUUUCAACA ...........((..(((((....(((((((((.((.((((.....)))-).)))))))))))((((.....))))...))))).-.)).(((.((((..-.)))).))).... ( -26.00) >consensus UUAAGUAAAAACGGGUGAGGAUGUGUAAUCGAUGUGGUUGCAAGUCGCA_GGUAAUCGAUUACUGUUGAAAGGACAAUGCCUCAAGGCGAAAACAGAAACAAUAUUAUUCAACA ........................(((((((((.....(((.....))).....))))))))).((((((........(((....)))(....).............)))))). (-20.20 = -20.36 + 0.16)

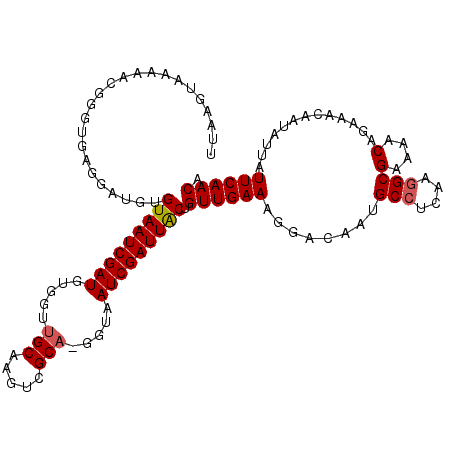
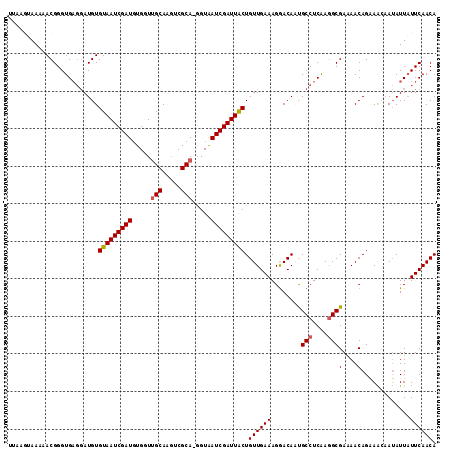
| Location | 11,406,329 – 11,406,443 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -18.60 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11406329 114 - 22407834 UGUUGAAUAAUAUUGUUUCUGUUUUCGCCUUGAGGCAGUGUCCUUUCAACAGCAAUCGAUUACUUUGCGACUUGCAAGCACAUCGAUUACACAUCCUCACCCGUUUUUAGUUAA (((((((..((((((((((............))))))))))...)))))))(.(((((((...(((((.....)))))...))))))).)........................ ( -25.90) >DroSec_CAF1 123808 113 - 1 UGUUGAAUAAUAUUGUUUCUGUUUUCGCCUUGAGGCAAUGUCCUUUCAACAGUAAUCGAUUACC-UGCGACUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUAU (((((((..((((((((((............))))))))))...)))))))(((((((((....-(((.....))).....)))))))))........................ ( -28.10) >DroSim_CAF1 130539 113 - 1 UGUUGAAUAAUAUUUUUUCUGUUUUCGCCUUGAGGCAUUGUCCUUUCAACAGUAAUCGAUUACC-UGCGAAUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUCA (((((((.............(....)(((....)))........)))))))(((((((((....-(((.....))).....)))))))))........................ ( -21.40) >DroEre_CAF1 131400 109 - 1 UGUUGAAUAAUUU-GCUACUGUUUUCGCCUCGAGGCGUUGUUCUUUCAACAGCAAUCGAUUACC-UGCGACUUGCCACCAAAUCGAUUACACAU---CACCCGUUUUUCCUCAG ((..(((.(((..-(((.........(((....)))((((......)))))))((((((((...-.((.....)).....))))))))......---.....))).)))..)). ( -19.80) >DroYak_CAF1 128013 111 - 1 UGUUGAAAAAUAU-UUUACUGUUCUCGC-UUGAGGCAUUGUCCUUUCAACAGUAAUCGAUCACC-UGCGACUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUCUUUAA ((((((((.....-..((.(((.(((..-..)))))).))...))))))))(((((((((....-(((.....))).....)))))))))........................ ( -20.60) >consensus UGUUGAAUAAUAUUGUUUCUGUUUUCGCCUUGAGGCAUUGUCCUUUCAACAGUAAUCGAUUACC_UGCGACUUGCAACCACAUCGAUUACACAUCCUCACCCGUUUUUACUUAA (((((((.............(....)(((....)))........)))))))(((((((((.....(((.....))).....)))))))))........................ (-18.60 = -19.40 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:16 2006