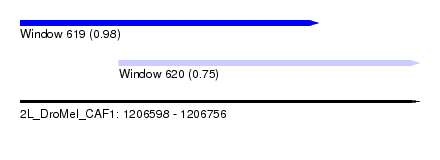
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,206,598 – 1,206,756 |
| Length | 158 |
| Max. P | 0.978660 |

| Location | 1,206,598 – 1,206,716 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -34.54 |
| Energy contribution | -35.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1206598 118 + 22407834 GGUCGGCUGCUUUUGGCCUU-UUCCCCCGGUGUUUGUCUUGGCCACUGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACA ....((((((..........-......((((.((..(((.(((....))))))..)).)))).((((...(((((.((((((((.....))))))))))))))))))).))))...... ( -39.70) >DroSec_CAF1 2311 118 + 1 GGUCGCCUGCUUUUGGCCUU-UUCCCCCGGUGUUUGUCUUGGCCACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACA ............(((((...-......((((.((..(((.(((....))))))..)).))))...((((((....(((((((((.....))))))))).....)))))).))))).... ( -39.70) >DroSim_CAF1 2302 118 + 1 GGUCGCCUGCCUUUGGCCUU-UUUCCCCGGUGUUUGUCUUGGCCACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACA (((.....))).(((((...-......((((.((..(((.(((....))))))..)).))))...((((((....(((((((((.....))))))))).....)))))).))))).... ( -41.10) >DroEre_CAF1 2236 119 + 1 GCUCGGCUGCUUUUGGCCCUUUUCCCUCGCUGUUUGUCUUGGCCACGGCCAGAUGAGAGCCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUUCGGGCUGGCCAAUACA ....((((......(((((....((((((((.((..(((.(((....))))))..)))).)))).))...((...(((((((((.....)))))))))....)))))))))))...... ( -45.10) >DroYak_CAF1 2387 119 + 1 GGUCGGCUGCUUUUGGCCUUUUUCCCCCGCUGUUUGUCUUGGCCACGGCCAGCUGAGUGCCGAGCAGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUUCGGGCUGGCCAAUACA ((((((((......)))).......(((((((((((...((((....))))((.....))))))))))..((...(((((((((.....)))))))))....)))))..))))...... ( -44.30) >consensus GGUCGGCUGCUUUUGGCCUU_UUCCCCCGGUGUUUGUCUUGGCCACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACA ............(((((..........((((.(((((((.(((....)))))))))).))))...((((((....(((((((((.....))))))))).....)))))).))))).... (-34.54 = -35.14 + 0.60)

| Location | 1,206,637 – 1,206,756 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1206637 119 + 22407834 GGC-CACUGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACAUAAUGCAAUAGAUAUGGCCAAAUGGCUGCUAAGAAGGCAG ...-..(((((...........(.((((((((....(((((((((.....))))))))).....)))))).)))..............(((....((((....)))).)))....))))) ( -32.60) >DroSec_CAF1 2350 119 + 1 GGC-CACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACAUACUGCAAUAGCUAUGGCCAAAUGGCUGCUAAGAAGGCAG (((-((.(((((....((......(((((((((((((((((((((.....)))))))))......((....)))))).)))))))).....)).)))))...)))))(((.....))).. ( -41.40) >DroSim_CAF1 2341 119 + 1 GGC-CACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGUGCUAGCCAAUACAUAAUGCAAUAGAUAUGGCCAAAUGGCUGCUAAGAAGGCAG (((-(((((...........))).(((.(((((((((((((((((.....)))))))))......((....)))))).)))).)))........)))))......(((((.....))))) ( -33.90) >DroEre_CAF1 2276 97 + 1 GGC-CACGGCCAGAUGAGAGCCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUUCGGGCUGGCCAAUACAUAAUGCAAUAGC----------------------AGGCAG .((-(..((((((.((((((((....))).......(((((((((.....)))))))))..)))))..)))))).........(((....))----------------------)))).. ( -37.10) >DroYak_CAF1 2427 119 + 1 GGC-CACGGCCAGCUGAGUGCCGAGCAGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUUCGGGCUGGCCAAUACAUAAUGCAAUAGCUGUGGGGCAAUGGCUGCUAAAAAGGCAG .((-(..((((((((.(.(((...))).).(((...(((((((((.....)))))))))....)))))))))))....((((..((....)))))).))).....(((((.....))))) ( -42.60) >DroPer_CAF1 2767 110 + 1 GGCCCACGUGCGGAUGC-------GUGAUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUUUAGGCUGUCCAUUGUU---UGCGAGUGCCAGAGCGGGACGACUAGUGUGUGGCCAG ((((.(((..(((((((-------.(((...(((((.((((((((.....)))))))))))))))).)).))))......---.....((.((.....)).)).....)..))))))).. ( -38.10) >consensus GGC_CACGGCCAGAUGAGAACCGAGCGGUAUGAUUGAUGUUGCAACAAUUUUGCAGCAUAAUUCCGGGCUAGCCAAUACAUAAUGCAAUAGCUAUGGCCAAAUGGCUGCUAAGAAGGCAG ........(((.............(((.(((((((((((((((((.....)))))))))........(....))))).)))).))).........((........))........))).. (-17.35 = -17.47 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:47 2006