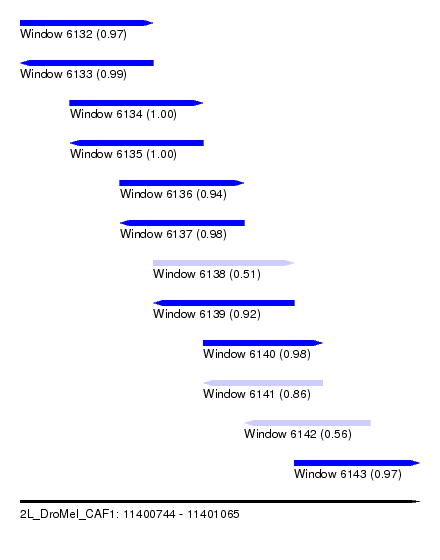
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,400,744 – 11,401,065 |
| Length | 321 |
| Max. P | 0.999807 |

| Location | 11,400,744 – 11,400,851 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400744 107 + 22407834 UAAAUACCUUUUAGGUAUCGUGCGUCUUCGGCACUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAGCAG ...(((((.....))))).(((.((....((((((((((..(((((((((.......)))))))..))...(((((....)))))))))).))))))).)))..... ( -37.20) >DroSec_CAF1 118230 107 + 1 UAAAUAACUUUUAUGUAUCGUGCGUCUUCGGCACUUGUUGCGGAUCGCUUCUUACAAAGGCGAUAACCAAGGUUGCAGCUGCAACAACAACGUGCCACACACAGCAG ...................(((.((....(((((((((((((((((((((.......)))))))(((....)))....)))))))))....))))))).)))..... ( -31.20) >DroSim_CAF1 125000 98 + 1 UAAAUAACUUUUAGGUAUCGUGCGUCUUCGGCACUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCAC--------- .............(((((.((((.......))))..((((.(((((((((.......)))))))..))...(((((....)))))..)))))))))..--------- ( -30.70) >DroEre_CAF1 125554 107 + 1 UAAAUAACUUUUGGGUAUAAUGCGUCUUCGACACUUGUUGCGGAUCGCCACUUACAACGGCGAUAACCAAGGCUGCAGCAGCAACAACAACGUGCCACACGCAGCAG ....................(((((....(.(((((((((.((((((((.........))))))..))...((((...))))..)))))).))))...))))).... ( -27.30) >DroYak_CAF1 121973 107 + 1 UAAAUAACUUUUAGGCAUAAUGCGUCCUCGACACUUGUUGCGGAUCGCCACUUACAACGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAACAG .............(((((.....(((...)))....((((.((((((((.........))))))..))...(((((....)))))..)))))))))........... ( -28.00) >consensus UAAAUAACUUUUAGGUAUCGUGCGUCUUCGGCACUUGUUGCGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAGCAG .............................((((((((((..((((((((.........))))))..))...(((((....)))))))))).)))))........... (-25.18 = -25.62 + 0.44)

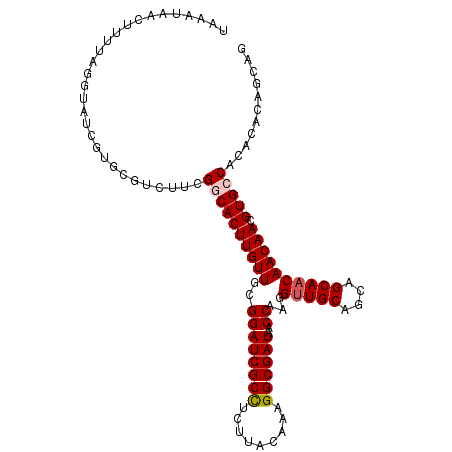
| Location | 11,400,744 – 11,400,851 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400744 107 - 22407834 CUGCUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGGUAUUUA .....(((((((((((.((((((((((....))).....((..((((((((.....))))))))))))))))))))))....))))))((((((.....)))))).. ( -43.60) >DroSec_CAF1 118230 107 - 1 CUGCUGUGUGUGGCACGUUGUUGUUGCAGCUGCAACCUUGGUUAUCGCCUUUGUAAGAAGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACAUAAAAGUUAUUUA ....((((((((((((.((((((((((....))).....((..(((((.(((....))))))))))))))))))))))....))))))).................. ( -32.30) >DroSim_CAF1 125000 98 - 1 ---------GUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGUUAUUUA ---------.((((((.((((((((((....))).....((..((((((((.....)))))))))))))))))))))))............................ ( -32.30) >DroEre_CAF1 125554 107 - 1 CUGCUGCGUGUGGCACGUUGUUGUUGCUGCUGCAGCCUUGGUUAUCGCCGUUGUAAGUGGCGAUCCGCAACAAGUGUCGAAGACGCAUUAUACCCAAAAGUUAUUUA ....(((((.((((((.((((((((((..(((((((...(((....)))))))).))..)))....)))))))))))))...))))).................... ( -34.70) >DroYak_CAF1 121973 107 - 1 CUGUUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCGUUGUAAGUGGCGAUCCGCAACAAGUGUCGAGGACGCAUUAUGCCUAAAAGUUAUUUA ....(((((.((((((.((((((((((..(((((((...(((....)))))))).))..)))....)))))))))))))...))))).................... ( -32.90) >consensus CUGCUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCGCAACAAGUGCCGAAGACGCACGAUACCUAAAAGUUAUUUA ....(((((.((((((.((((((((((....))).....((..((((((((.....)))))))))))))))))))))))...))))).................... (-31.86 = -32.22 + 0.36)

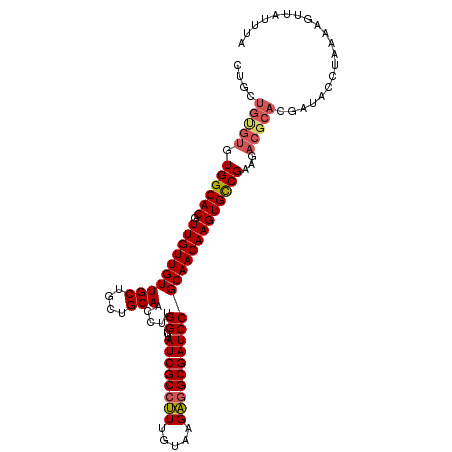
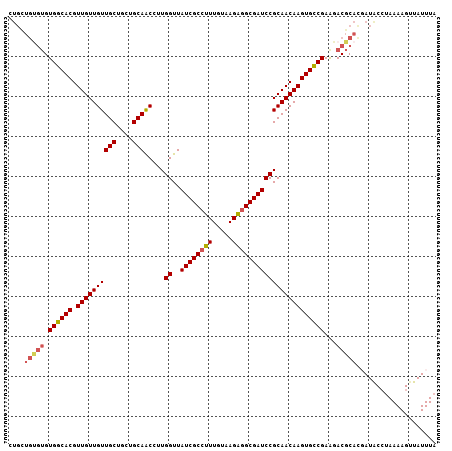
| Location | 11,400,784 – 11,400,891 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.13 |
| SVM RNA-class probability | 0.999807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400784 107 + 22407834 CGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAGCAGCAACAUGGACAACAACAAAUAACAUUGUCUGAAAGUUGUU .(((((((((.......)))))))..))...(((((....)))))(((((((((.....)))...........((((((............))))))....)))))) ( -27.90) >DroSec_CAF1 118270 107 + 1 CGGAUCGCUUCUUACAAAGGCGAUAACCAAGGUUGCAGCUGCAACAACAACGUGCCACACACAGCAGCAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUU .(((((((((.......)))))))..))...(((((....)))))(((((((((.....)))..........(((((((............)))))))...)))))) ( -28.10) >DroSim_CAF1 125040 94 + 1 CGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCAC-------------ACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUU .(((((((((.......)))))))..))...(((((....)))))(((((((.....)-------------.(((((((............)))))))...)))))) ( -27.00) >DroEre_CAF1 125594 106 + 1 CGGAUCGCCACUUACAACGGCGAUAACCAAGGCUGCAGCAGCAACAACAACGUGCCACACGCAGCAGCAACACGG-CAACAACAAAUAACAUUGUCUGAAAGUUGUU .((((((((.........))))))..))...(((((....(((.(......)))).....)))))((((((..(.-...).((((......))))......)))))) ( -26.20) >DroYak_CAF1 122013 107 + 1 CGGAUCGCCACUUACAACGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAACAGCAACACGGACAACAACAAAUAACAUUGUCUGAAUGUUGUU .((((((((.........))))))..))...(((((....)))))......(((.....)))(((((((...(((((((............)))))))..))))))) ( -29.40) >consensus CGGAUCGCCUCUUACAAAGGCGAUAACCAAGGUUGCAGCAGCAACAACAACGUGCCACACACAGCAGCAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUU .((((((((.........))))))..))...(((((....)))))(((((((((.....)))..........(((((((............)))))))...)))))) (-24.58 = -25.14 + 0.56)

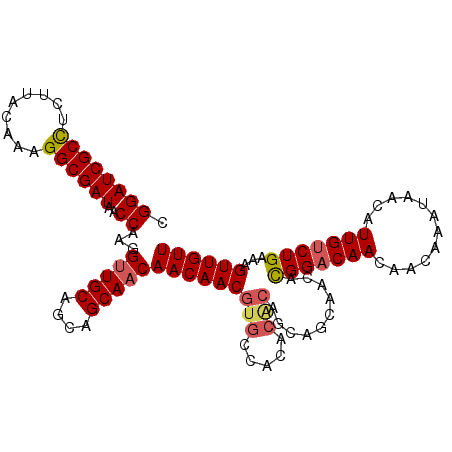
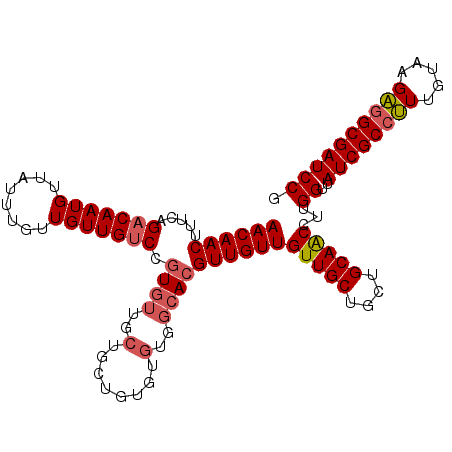
| Location | 11,400,784 – 11,400,891 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -30.66 |
| Energy contribution | -31.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400784 107 - 22407834 AACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCAUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCG ((((((.....(((((((........))))))).....(((..(.....)..))).))))))(((((....)))))...((..((((((((.....)))))))))). ( -35.60) >DroSec_CAF1 118270 107 - 1 AACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGCAGCUGCAACCUUGGUUAUCGCCUUUGUAAGAAGCGAUCCG ((((((.....(((((((........))))))).((((..(........)..))))))))))(((((....)))))...((..(((((.(((....)))))))))). ( -32.00) >DroSim_CAF1 125040 94 - 1 AACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGU-------------GUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCG ((((((.....(((((((........))))))).((-------------(...)))))))))(((((....)))))...((..((((((((.....)))))))))). ( -29.70) >DroEre_CAF1 125594 106 - 1 AACAACUUUCAGACAAUGUUAUUUGUUGUUG-CCGUGUUGCUGCUGCGUGUGGCACGUUGUUGUUGCUGCUGCAGCCUUGGUUAUCGCCGUUGUAAGUGGCGAUCCG ..((((.....(((((((.....))))))).-....))))..((((((.(..(((((....)).)))..)))))))...((..(((((((.......))))))))). ( -34.70) >DroYak_CAF1 122013 107 - 1 AACAACAUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGUUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCGUUGUAAGUGGCGAUCCG ((((((.....(((((((........))))))).((((..(........)..))))))))))(((((....)))))...((..(((((((.......))))))))). ( -36.20) >consensus AACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGCUGCUGCAACCUUGGUUAUCGCCUUUGUAAGAGGCGAUCCG ((((((.....(((((((........))))))).((((..(........)..))))))))))(((((....)))))...((..((((((((.....)))))))))). (-30.66 = -31.22 + 0.56)

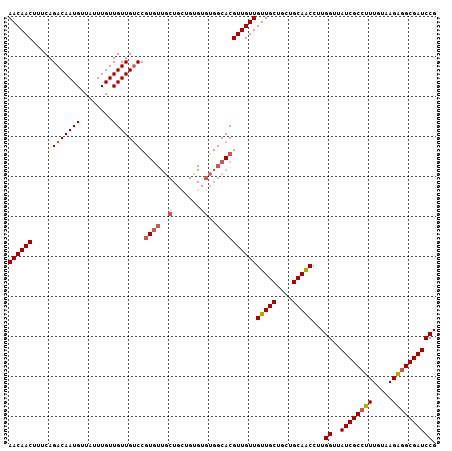
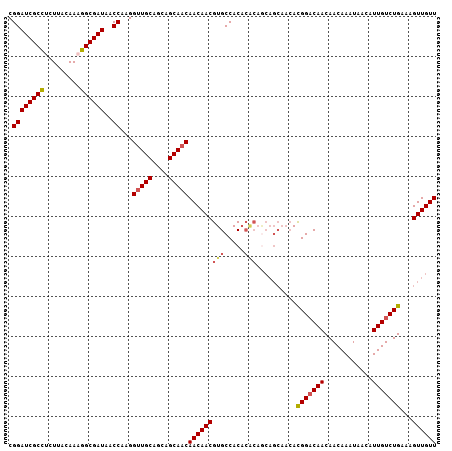
| Location | 11,400,824 – 11,400,924 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -19.44 |
| Energy contribution | -21.76 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400824 100 + 22407834 GCAACAACAACGUGCCACACACAGCAGCAACAUGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAACGAGCAACUGCAACAUCGGC ((........(((((.....(((((((((((.(((((((............)))))))...)))))))))))....)).))).((....)).......)) ( -25.90) >DroSec_CAF1 118310 100 + 1 GCAACAACAACGUGCCACACACAGCAGCAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAGCGAGCAACUGCAACAUCGGC .............(((....(((((((((((.(((((((............)))))))...)))))))))))...(((((.......))))).....))) ( -31.30) >DroSim_CAF1 125080 87 + 1 GCAACAACAACGUGCCAC-------------ACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUACUGUUCCUGCAGCGAGCAACUGCAACAUCGGC .............(((..-------------.(((((((............)))))))..(((((((.((((....))))..)))))))........))) ( -20.60) >DroEre_CAF1 125634 99 + 1 GCAACAACAACGUGCCACACGCAGCAGCAACACGG-CAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUGCCUGCAACGAGCAACUGCAACAUCGGC .............(((...((..((((...(((((-(((((((..................)))))))))))).))))..)).((....))......))) ( -29.37) >DroYak_CAF1 122053 100 + 1 GCAACAACAACGUGCCACACACAACAGCAACACGGACAACAACAAAUAACAUUGUCUGAAUGUUGUUGCUGUUCCUGCAAUGAGCAACUGCAACAUCGGC (((((((((..(((.....)))..........(((((((............)))))))..)))))))))(((...(((.....)))...)))........ ( -24.70) >consensus GCAACAACAACGUGCCACACACAGCAGCAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAACGAGCAACUGCAACAUCGGC .............(((....(((((((((((.(((((((............)))))))...)))))))))))...((((.........)))).....))) (-19.44 = -21.76 + 2.32)

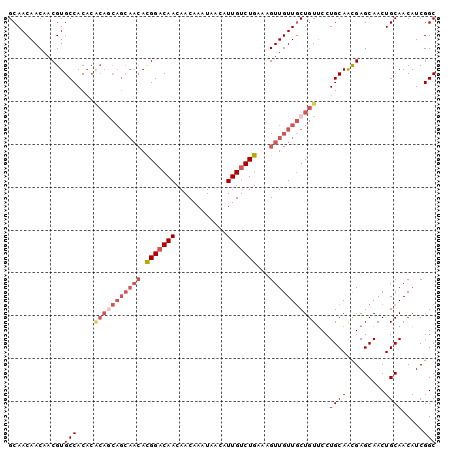
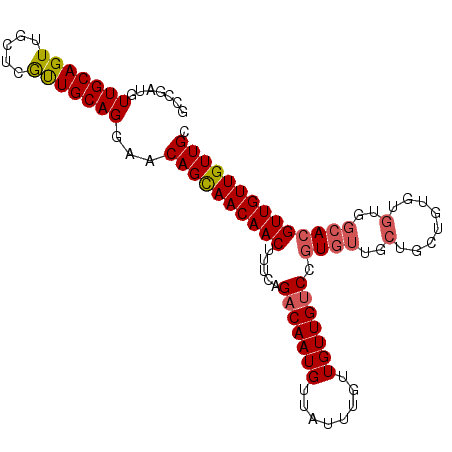
| Location | 11,400,824 – 11,400,924 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.34 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400824 100 - 22407834 GCCGAUGUUGCAGUUGCUCGUUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCAUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGC ((((....((((((.((.....((((((.(((((((((.(..((.....))..).)))))))))))).))).)).))))))..))))............. ( -31.10) >DroSec_CAF1 118310 100 - 1 GCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGC .....(.(((((((.....))))))).).((((((((((.....(((((((........))))))).((((..(........)..)))))))))))))). ( -36.00) >DroSim_CAF1 125080 87 - 1 GCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGUAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGU-------------GUGGCACGUUGUUGUUGC ...(.(.(((((((.....))))))).).).((((((((.....(((((((........)))))))(((-------------(...))))..)))))))) ( -24.30) >DroEre_CAF1 125634 99 - 1 GCCGAUGUUGCAGUUGCUCGUUGCAGGCACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUG-CCGUGUUGCUGCUGCGUGUGGCACGUUGUUGUUGC ((((((((.((((((((.....)))(((((.((((((((...((.....))......)))))))-).)))))))))).)))).))))............. ( -35.50) >DroYak_CAF1 122053 100 - 1 GCCGAUGUUGCAGUUGCUCAUUGCAGGAACAGCAACAACAUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGUUGUGUGUGGCACGUUGUUGUUGC .....(.(((((((.....))))))).).((((((((((.....(((((((........))))))).((((..(........)..)))))))))))))). ( -33.90) >consensus GCCGAUGUUGCAGUUGCUCGUUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUGCUGCUGUGUGUGGCACGUUGUUGUUGC .......(((((((.....)))))))...((((((((((.....(((((((........))))))).((((..(........)..)))))))))))))). (-26.90 = -27.34 + 0.44)

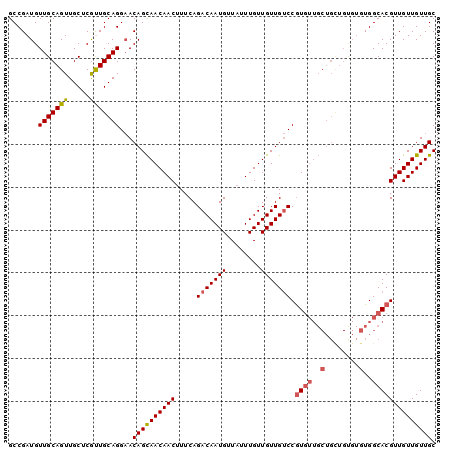
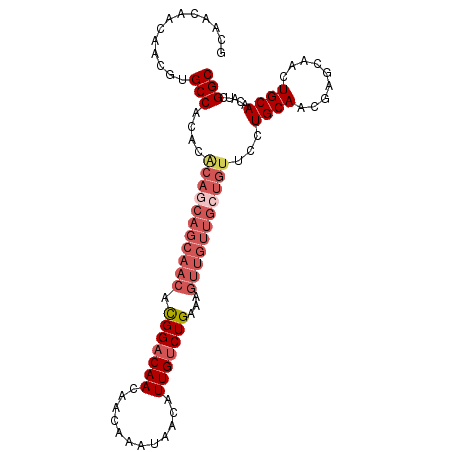
| Location | 11,400,851 – 11,400,964 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400851 113 + 22407834 CAACAUGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAACGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCG ......((((((((((((....((.....))....)))))).((((((.(((((..((....))......((.(((....)))))....)))))....)))))).)))).)). ( -27.50) >DroSec_CAF1 118337 113 + 1 CAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAGCGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCG ((((.(((((((............)))))))...)))).(((((...(((((((((((....))....)).)))))))))))).(((((((..((...))...)))))))... ( -32.40) >DroSim_CAF1 125098 109 + 1 ----ACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUACUGUUCCUGCAGCGAGCAACUGCAACAUCGGCUGCAGCAGUACCACAAGUUGCCACUGGAAUAUUUGUGCCG ----.(((((((............)))))))........(((((...(((((((((((....))....)).)))))))))))).(((((((..((...))...)))))))... ( -28.90) >DroEre_CAF1 125661 102 + 1 CAACACGG-CAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUGCCUGCAACGAGCAACUGCAACAUCGGCAGCAGCAGCAGCACAAGUUGCCACUGGAAU---------- ......((-((((.((((......))))......((((((((((((((((((.........)))).....)))).))))))))))....))))))........---------- ( -35.50) >DroYak_CAF1 122080 113 + 1 CAACACGGACAACAACAAAUAACAUUGUCUGAAUGUUGUUGCUGUUCCUGCAAUGAGCAACUGCAACAUCGGCAGCAGCAGCAACACAAGUUGCCACUGGAAUAUUUGUGCCG ((((((((((((............)))))))..)))))((((((((.((((.(((.((....))..)))..)))).))))))))(((((((..((...))...)))))))... ( -34.00) >consensus CAACACGGACAACAACAAAUAACAUUGUCUGAAAGUUGUUGCUGUUCCUGCAACGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCG ......((.((((.((((......))))......(.(((((((((..(((......((....)).....)))..))))))))).)....)))))).................. (-22.02 = -22.06 + 0.04)

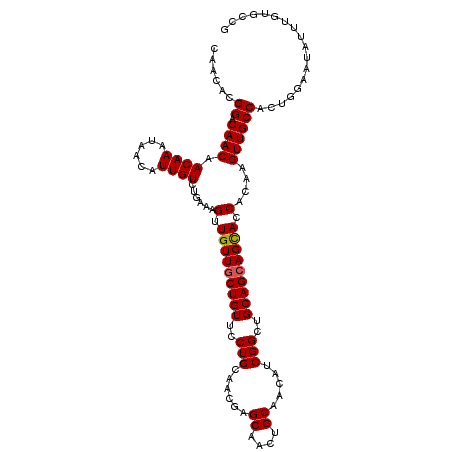
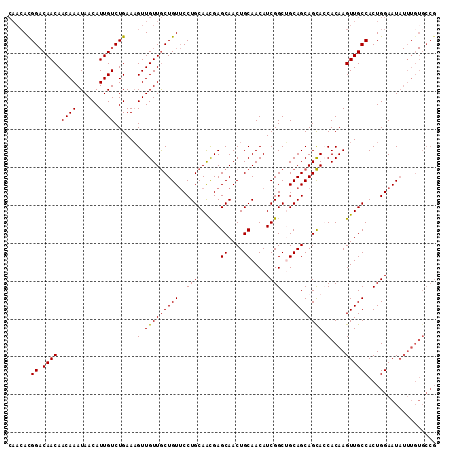
| Location | 11,400,851 – 11,400,964 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -28.40 |
| Energy contribution | -27.88 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400851 113 - 22407834 CGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGUUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCAUGUUG (((((..............(((((.....(.((.((((((((....)))))))).)).)))))).(((.(((((((((.(..((.....))..).)))))))))))).))))) ( -36.60) >DroSec_CAF1 118337 113 - 1 CGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUG ((((((..........(..((..(....)(.((.((((((((....)))))))).)).)))..).(((.(((((((((.(..((.....))..).)))))))))))))))))) ( -41.20) >DroSim_CAF1 125098 109 - 1 CGGCACAAAUAUUCCAGUGGCAACUUGUGGUACUGCUGCAGCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGUAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGU---- (((.((((...............((((..((...((((((((....)))))))).....))..))))((((((((((............))))).))))).))))))).---- ( -29.90) >DroEre_CAF1 125661 102 - 1 ----------AUUCCAGUGGCAACUUGUGCUGCUGCUGCUGCCGAUGUUGCAGUUGCUCGUUGCAGGCACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUG-CCGUGUUG ----------....(((..(((.(....).)))..)))((((.((((..((....)).))))))))((((.((((((((...((.....))......)))))))-).)))).. ( -34.60) >DroYak_CAF1 122080 113 - 1 CGGCACAAAUAUUCCAGUGGCAACUUGUGUUGCUGCUGCUGCCGAUGUUGCAGUUGCUCAUUGCAGGAACAGCAACAACAUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUG ((((((........(((..(((((....)))))..))).(((.((((..((....)).)))))))(((.(((((((((....((.....))....)))))))))))))))))) ( -41.10) >consensus CGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGUUGCAGGAACAGCAACAACUUUCAGACAAUGUUAUUUGUUGUUGUCCGUGUUG ..............(((((((((((.((..(((.((....)).).))..)))))))).)))))..(((.(((((((((....((.....))....))))))))))))...... (-28.40 = -27.88 + -0.52)

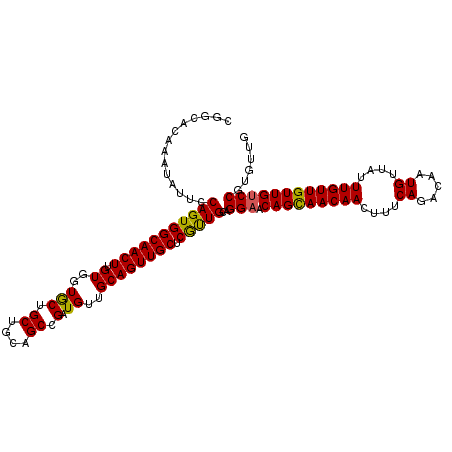
| Location | 11,400,891 – 11,400,987 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400891 96 + 22407834 GCUGUUCCUGCAACGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCGACUUUGCUG-----------------UGCAGCAACAUCCA ((.(((.((......)).))).)).......((((((.(((((....((((((.(((..........))).)))))))))))-----------------))))))........ ( -29.70) >DroSec_CAF1 118377 81 + 1 GCUGUUCCUGCAGCGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCGACUUCU-UG------------------------------- ((((...(((((((((((....))....)).))))))))))).....((((((.(((..........))).))))))..-..------------------------------- ( -25.00) >DroSim_CAF1 125134 109 + 1 ACUGUUCCUGCAGCGAGCAACUGCAACAUCGGCUGCAGCAGUACCACAAGUUGCCACUGGAAUAUUUGUGCCGACUUCU-UGUCGGCCGACCGA---CCAGCAGCAACAUCCA ((((...(((((((((((....))....)).))))))))))).......(((((..((((.....(((.((((((....-.)))))))))....---))))..)))))..... ( -40.80) >DroYak_CAF1 122120 112 + 1 GCUGUUCCUGCAAUGAGCAACUGCAACAUCGGCAGCAGCAGCAACACAAGUUGCCACUGGAAUAUUUGUGCCGACUACU-UGUCGGCCGACCAGCAGCAAGCAGCAACAUCCA ((((((.((((.....((..((((.......))))..)).(((((....)))))...(((.....(((.((((((....-.))))))))))))))))..))))))........ ( -40.30) >consensus GCUGUUCCUGCAACGAGCAACUGCAACAUCGGCUGCAGCAGCACCACAAGUUGCCACUGGAAUAUUUGUGCCGACUUCU_UG_________________AGCAGCAACAUCCA ((((...(((((((((((....))....)).))))))))))).....((((((.(((..........))).)))))).................................... (-20.04 = -20.47 + 0.44)

| Location | 11,400,891 – 11,400,987 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -22.56 |
| Energy contribution | -25.12 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400891 96 - 22407834 UGGAUGUUGCUGCA-----------------CAGCAAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGUUGCAGGAACAGC ....((((.(((((-----------------((((((...((((((.......(.(((....))).)..))))))(((((((....)))))))))))).).))))).)))).. ( -34.00) >DroSec_CAF1 118377 81 - 1 -------------------------------CA-AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGC -------------------------------..-.......(.(((..........))).)..((((..((((.((((((((....)))))))).))..))..))))...... ( -26.20) >DroSim_CAF1 125134 109 - 1 UGGAUGUUGCUGCUGG---UCGGUCGGCCGACA-AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUACUGCUGCAGCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGU ....((((((..((((---...((..((((((.-....))))))....))...))))..))..((((..((...((((((((....)))))))).....))..)))))))).. ( -46.10) >DroYak_CAF1 122120 112 - 1 UGGAUGUUGCUGCUUGCUGCUGGUCGGCCGACA-AGUAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGUUGCUGCUGCUGCCGAUGUUGCAGUUGCUCAUUGCAGGAACAGC ....((((.((((..(((((..((((((((((.-....))))))..........(((..(((((....)))))..)))....))))...)))))........)))).)))).. ( -46.20) >consensus UGGAUGUUGCUGCU_________________CA_AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCAGCCGAUGUUGCAGUUGCUCGCUGCAGGAACAGC ....((((.((((.......................((((.(.(((..........))).).))))((((.((.((((((((....)))))))).)))))).)))).)))).. (-22.56 = -25.12 + 2.56)

| Location | 11,400,924 – 11,401,025 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.42 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -13.01 |
| Energy contribution | -15.17 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400924 101 - 22407834 CCGAAACUGGCCAUUGCCACUUCAUCAUAGGCCAGGUUUGGAUGUUGCUGCA-----------------CAGCAAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCA (((((.((((((.................)))))).))))).(((.((.(((-----------------(.((((.((.(.(((..........))).).)))))).)))).)).))) ( -34.73) >DroSec_CAF1 118410 76 - 1 CCGAAACUGGCCAUUGCCACUUCAUCAU-----------------------------------------CA-AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCA ((((...((((....))))((((.....-----------------------------------------..-.))))))))...........(((..(((.(....).)))..))).. ( -21.00) >DroSim_CAF1 125167 114 - 1 CCGAAACUGGCCAUUGCCACUUCAUCAUAGGCCAGGUUUGGAUGUUGCUGCUGG---UCGGUCGGCCGACA-AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUACUGCUGCA (((((.((((((.................)))))).)))))..(((((..((((---...((..((((((.-....))))))....))...))))..))))).((..(.....)..)) ( -47.53) >DroYak_CAF1 122153 117 - 1 CCGAAACUGACCAUUGCCACUUCAUCAUAGGCCAGGUUUGGAUGUUGCUGCUUGCUGCUGGUCGGCCGACA-AGUAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGUUGCUGCUGCU ........((((...(((...........)))..)))).(((((((..((((.((((((.(((....))).-)))))).))))..)))))))(((..(((((....)))))..))).. ( -41.80) >DroAna_CAF1 106502 106 - 1 CUUU--GUGCCCAUUGCCACUUCAUCAUAGGCCGGGUUUGGAUGUUGCUAC-------UGGACGGUCAGCC-UUAAGUCGCCAAAAAUAUUCCGGUGGCAACUUGUGCCGAUGCUG-- ....--..((((...(((...........))).))))((((..((((((((-------((((.((....))-....(....)........)))))))))))).....)))).....-- ( -29.60) >consensus CCGAAACUGGCCAUUGCCACUUCAUCAUAGGCCAGGUUUGGAUGUUGCUGC________GG_CGG_C__CA_AGAAGUCGGCACAAAUAUUCCAGUGGCAACUUGUGGUGCUGCUGCA ..............(((((((((......((((..............................))))......))))).)))).........((((((((.(....).)))))))).. (-13.01 = -15.17 + 2.16)

| Location | 11,400,964 – 11,401,065 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -26.45 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11400964 101 + 22407834 ACUUUGCUG-----------------UGCAGCAACAUCCAAACCUGGCCUAUGAUGAAGUGGCAAUGGCCAGUUUCGGACCAGACACCGAUAUUGGCCAACACGGUAGCUGACCAGCA ....(((((-----------------..((((.((...........(((...........)))..((((((((.((((........)))).)))))))).....)).))))..))))) ( -38.00) >DroSim_CAF1 125207 111 + 1 ACUUCU-UGUCGGCCGACCGA---CCAGCAGCAACAUCCAAACCUGGCCUAUGAUGAAGUGGCAAUGGCCAGUUUCGGCCCAGACACCGAUAUUGGCCAACACGGUAGCUGACCA--- ......-.((((((..((((.---...((.((..((((..............))))..)).))..((((((((.((((........)))).))))))))...)))).))))))..--- ( -39.44) >DroEre_CAF1 125763 104 + 1 ---ACU-UGUCGGCCGAGCAGCA-------GCUACAUCCAAACCUGGCCUAUGAUGAAGUGGCAUUGGUCAGUUUCGGCCCAGACACCGAUAUUGGCCAACUCGAUAGCCGACCG--- ---...-.((((((((((..((.-------(((.((((..............)))).))).)).(((((((((.((((........)))).)))))))))))))...))))))..--- ( -38.54) >DroYak_CAF1 122193 114 + 1 ACUACU-UGUCGGCCGACCAGCAGCAAGCAGCAACAUCCAAACCUGGCCUAUGAUGAAGUGGCAAUGGUCAGUUUCGGCCCAGACACCGAUAUUGGCCAACUCGGUAGCUGACCA--- ......-.((((((..(((.((.((...((.((....(((....)))....)).))..)).))..((((((((.((((........)))).))))))))....))).))))))..--- ( -34.90) >consensus ACUACU_UGUCGGCCGACCAG_____AGCAGCAACAUCCAAACCUGGCCUAUGAUGAAGUGGCAAUGGCCAGUUUCGGCCCAGACACCGAUAUUGGCCAACACGGUAGCUGACCA___ ........((((((................................(((...........)))..((((((((.((((........)))).))))))))........))))))..... (-26.45 = -27.32 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:12 2006