
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,398,779 – 11,398,944 |
| Length | 165 |
| Max. P | 0.558719 |

| Location | 11,398,779 – 11,398,881 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -19.94 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
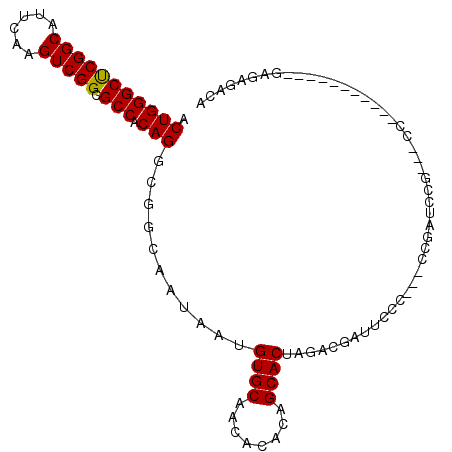
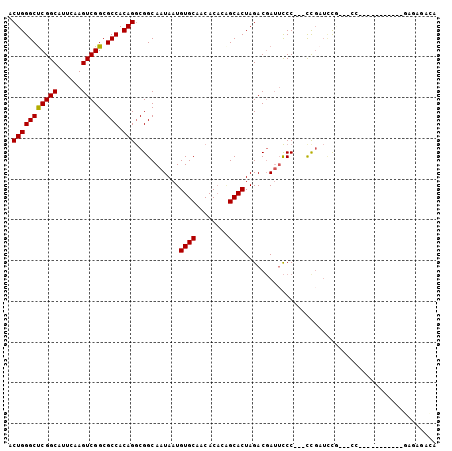
>2L_DroMel_CAF1 11398779 102 - 22407834 ACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACAGAGCACUAGACGAUUCCC---UUCAUUUAGGCCCCCCCUUCCAACGAGAGACA ...(((...(((......((((.((((...)))).)......((((........))))..)))((.....---.))......)))..)))......(....)... ( -25.70) >DroSec_CAF1 115608 101 - 1 ACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCCC---CCGAUCCG-AACCCCUUUCCCUACUAGAGCCA ....(((((((.......((((.((((...)))).)......((((........))))..))).......---)).....(-((.....))).......))))). ( -26.94) >DroSim_CAF1 123035 84 - 1 ACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACUGCACUAGACGAUUCCC---CCGAUCCG-AACC-----------------CA ...(((.((((.......(((((((((...))))........(((((......)))))............---))))))))-).))-----------------). ( -26.81) >DroEre_CAF1 123610 86 - 1 ACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACGGCACUAGACGAUUCCC---CCGAUCCG----------------GAGAGUCG .(((((.((((.......((((.((((...)))).)......((((........))))..))).......---))))))))----------------)....... ( -27.94) >DroYak_CAF1 119796 88 - 1 ACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCCCACUCCGCUAC-----------------GAAAGUCA .(((((((((((......))))).))).)))((((.......((((........))))..((....)).....)))).((-----------------....)).. ( -25.10) >DroAna_CAF1 104408 96 - 1 ACUGGGCCCGGCAUUCGAGUCGGCGCCACAGACGGCAAUAAUGUGCAGCACCCAGCACUAGACCAGAUCC---CCGAACUU-CCCC-----CAAAGCUGGCGAAA .(((((((((((......))))).))).))).((((......((((........)))).......((...---.......)-)...-----....))))...... ( -26.10) >consensus ACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAUAAUGUGCAACACACAGCACUAGACGAUUCCC___CCGAUCCG___CC___________GAGAGACA .(((((((((((......))))).))).)))...........((((........))))............................................... (-19.94 = -19.80 + -0.14)

| Location | 11,398,842 – 11,398,944 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.58 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -25.80 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
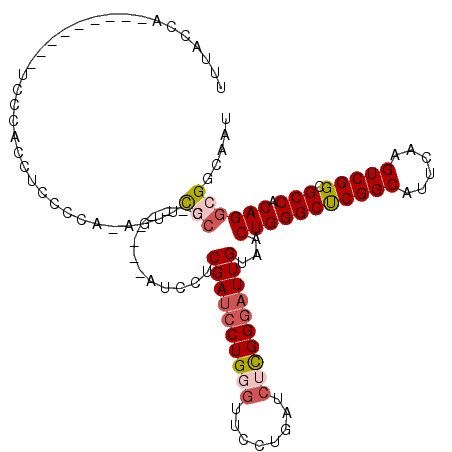
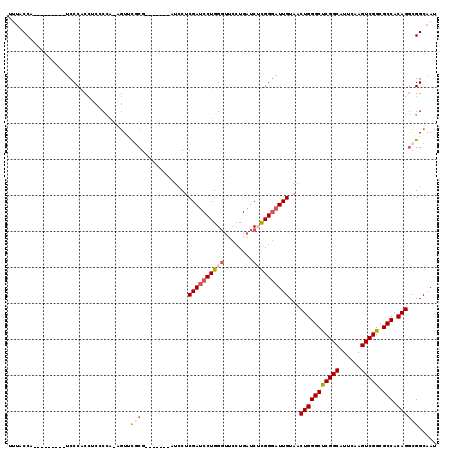
>2L_DroMel_CAF1 11398842 102 - 22407834 UUUACCA---------UCCCACCCCACCA-AGUUCGCG-------AUCCUCGAUCCUGGGUUCCUGAUCUCGGGAUUGUAACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAU .......---------.............-.((.(((.-------.....((((((((((........))))))))))...(((((((((((......))))).))).))))))))... ( -32.70) >DroSec_CAF1 115670 102 - 1 UUUACCA---------UCCCACCUCCCCA-AGUUCGCG-------AUCCUCGAUCCUGGGUUCCUGAUCGCGGGAUUGUAACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAU .......---------..((.(((.....-((((.(((-------((((((((((..((...)).))))).)))))))))))).((((((((......))))).)))..))).)).... ( -35.10) >DroSim_CAF1 123080 102 - 1 UUUACCA---------UCCCACCUUCCCA-AGUUCGCG-------AUCCUCGAUCCUGGGUUCCUGAUCGCGGGAUUGUAACUGGGCUCGGCAUUUAAGUCGGCGCCACAGGCGGCAAU .......---------..((.(((.....-((((.(((-------((((((((((..((...)).))))).)))))))))))).((((((((......))))).)))..))).)).... ( -35.10) >DroEre_CAF1 123657 102 - 1 CUCGCCA---------UCCCACCUCCCCA-AGUGCGCG-------AUCCUCGAUCCUGGGUGCCUGAUCUCGGGAUUGUAACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAU ...(((.---------...(((.......-.))).((.-------.....((((((((((........))))))))))...(((((((((((......))))).))).))))))))... ( -36.70) >DroYak_CAF1 119845 119 - 1 CUUGCCACUUGCCACUUCCCACCUCGCCAAAGUACUCGAUUCUCGAUACUCGAUCCUGGGUUCCUGAUCUCGGGAUUGUAACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAU .(((((....(((((((....((..(((..((((.(((.....)))))))((((((((((........))))))))))......)))..)).....)))).)))(((...)))))))). ( -41.60) >DroAna_CAF1 104465 101 - 1 UUUACCU-------CUACCCGCA-GUCUG-GCUUUUCG-------GCUCUCGAUCCUGGCUA--UGUUCUUGGCUUUGUAACUGGGCCCGGCAUUCGAGUCGGCGCCACAGACGGCAAU .......-------......((.-(((((-.....(((-------((((..(((((.((((.--.((((........).)))..)))).)).))).))))))).....))))).))... ( -30.30) >consensus UUUACCA_________UCCCACCUCCCCA_AGUUCGCG_______AUCCUCGAUCCUGGGUUCCUGAUCUCGGGAUUGUAACUGGGCUCGGCAUUCAAGUCGGCGCCACAGGCGGCAAU ..................................(((.............((((((((((........))))))))))...(((((((((((......))))).))).))))))..... (-25.80 = -26.72 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:00 2006