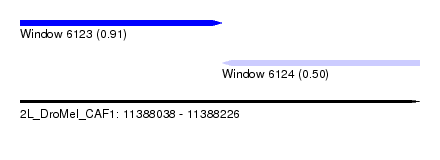
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,388,038 – 11,388,226 |
| Length | 188 |
| Max. P | 0.909104 |

| Location | 11,388,038 – 11,388,133 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11388038 95 + 22407834 CUAUUUAAGGCCAAGAUUUGAUUGUCAAGCGGAGAUAUGCCAAUUUCUCAGUCG-------GAUCCUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((.(((...(((((...(((.....)-------))...))))).)))...))))).))).))..))..))))). ( -32.40) >DroPse_CAF1 130374 95 + 1 CUAUUUAAGGCCGAGAUUUGAUUGUCAAGCGGAGAUAUGCCAAUUUCUCAGUCG-------AGUCUUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((.(((...((((((...(((....)-------))..)))))).)))...))))).))).))..))..))))). ( -34.40) >DroYak_CAF1 108589 95 + 1 CUAUUUAAGGCCAAGAUUUGAUUGUCAAGCGGAGAUAUGCCAAUUUCUCAGUCG-------GAUCCUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((.(((...(((((...(((.....)-------))...))))).)))...))))).))).))..))..))))). ( -32.40) >DroMoj_CAF1 141126 102 + 1 CUAUUUAAGGCCGAGAUUUGAUUGUCAAGCGUAGAUAUGCCAAUUUCUCUGUCGCCAUUUGGGCCUUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((((.((..((((((..........(((.....))).))))))..)))).))))).))).))..))..))))). ( -29.00) >DroAna_CAF1 94599 95 + 1 CUAUUUAAGGCCGAGAUUUGAUUGUCCGGCGGAGAUAUGCCAAUUUCUCAGUCG-------GUUCCUGGCAGCUCACAGCUUGUCGGACAUUGUUCUAGCUA ........(((..(((......(((((((((((((.((....)).)))).((((-------(...)))))(((.....))))))))))))....))).))). ( -27.10) >DroPer_CAF1 129480 95 + 1 CUAUUUAAGGCCGAGAUUUGAUUGUCAAGCGGAGAUAUGCCAAUUUCUCAGUCG-------AGUCUUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((.(((...((((((...(((....)-------))..)))))).)))...))))).))).))..))..))))). ( -34.40) >consensus CUAUUUAAGGCCGAGAUUUGAUUGUCAAGCGGAGAUAUGCCAAUUUCUCAGUCG_______GGUCCUGGCAGCUCACAGCUUGUCGGACAUUGUUCUGGCUA ........(((((..((.((.(((.(((((.(((...(((((........................))))).)))...))))).))).))..))..))))). (-26.44 = -26.58 + 0.14)


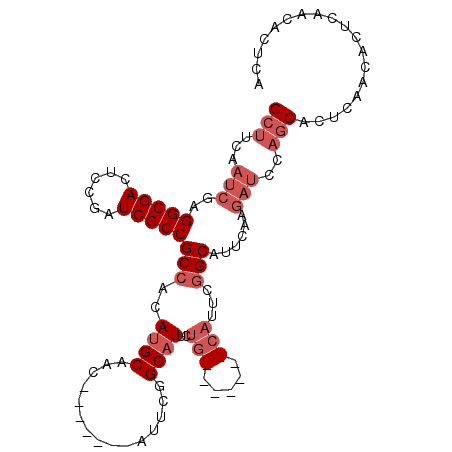
| Location | 11,388,133 – 11,388,226 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -12.24 |
| Energy contribution | -13.91 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11388133 93 - 22407834 GCUUCAAUCGAGGCGACUCCAAUCGCCGCCACAUGCAAC-------AUUCGGCAUUCUG-------CAUUCGGCAUUCAAGAUCCAGCACUCAACACUCAACACUCA (((...(((..(((((......)))))(((..(((((..-------...........))-------)))..)))......)))..)))................... ( -21.12) >DroSec_CAF1 105094 93 - 1 GCUUCAAUCGAGGCGACUCCGAUCGCCGCCACAUGCAAC-------AUUCGGCAUUCUG-------CAUUCGGCAUCCAAGAUCCAGCACUCAACACUCAACACUCA (((...(((..(((((......)))))(((..(((((..-------...........))-------)))..)))......)))..)))................... ( -21.12) >DroSim_CAF1 112032 86 - 1 GCUUCAAUCGAGGCGACUCCGAUCGCCGCCACAUGCAAC-------AUUCGGCUUUCUG-------CAUUCGGCAUCCAAGAUCCAGCACUCA-------ACACUCA (((...(((..(((((......)))))(((..(((((..-------...........))-------)))..)))......)))..))).....-------....... ( -21.12) >DroEre_CAF1 108699 100 - 1 GCUUCAAUCGAGGCGACUCCGAUCGCCGCCACAUGCAAC-------AUUCGGCAUUCUGCAUUCGGCAUUCGGCAUUCAAGAUCCAGCACUCAACACUCAACACUCA (((...(((..(((((......)))))(((..((((...-------.....((.....)).....))))..)))......)))..)))................... ( -22.82) >DroYak_CAF1 108684 107 - 1 GCUUCAAUCGAGGCGACUCCGAUCGCCGCCACAUGCAACAUUCAACAUUCGGCAUUCUGCAUUCUGCAUUCGGCAUUCAAGAUCCAGCACUCAACACUCAACACUCA (((...(((..(((((......)))))(((..(((((........(....)((.....))....)))))..)))......)))..)))................... ( -22.20) >DroAna_CAF1 94694 82 - 1 GCAUCAAUCGAGGCGACUGCGAUCGCCGCCGCAUGCAAC-------AUU--GCAUCGUA----------GCAGCAUA------GCAACUUGCAACACCCAACACUCA ((.........(((((......)))))((..((((((..-------..)--)))).)..----------)).))...------((.....))............... ( -21.00) >consensus GCUUCAAUCGAGGCGACUCCGAUCGCCGCCACAUGCAAC_______AUUCGGCAUUCUG_______CAUUCGGCAUUCAAGAUCCAGCACUCAACACUCAACACUCA (((...(((..(((((......)))))(((..((((...............))))..(((.....)))...)))......)))..)))................... (-12.24 = -13.91 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:54 2006