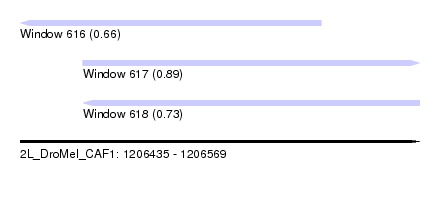
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,206,435 – 1,206,569 |
| Length | 134 |
| Max. P | 0.889027 |

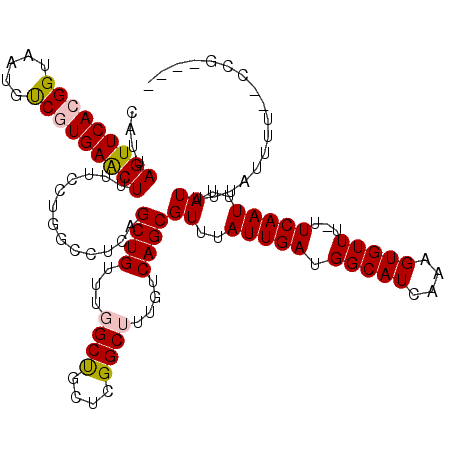
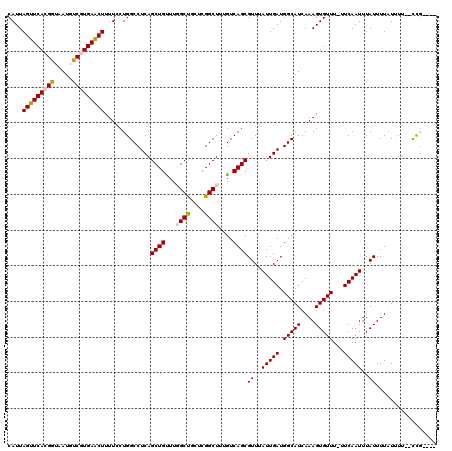
| Location | 1,206,435 – 1,206,536 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1206435 101 - 22407834 GCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUGUUGAUAC-------------------UGGGCACGACCUCU ((((((((((....(((.....(((...((((......)))).))))))....((((((((.......))))))))..)))))))..-------------------..)))......... ( -30.70) >DroPse_CAF1 2574 107 - 1 GCCAUCAAUAAACGCUGACAGAGCCAA-UGGCAGAGCAGCUGAGGCCAG-AAGAGCUCAUGGCAUUACCGUGAACUAAUGUUGAUACGGGGGC----------CACCGGGCACGACCCC- ((((((((((...((((.....(((..-.)))....)))).........-...((.((((((.....)))))).))..))))))).(((....----------..))))))........- ( -31.40) >DroSim_CAF1 2139 101 - 1 GCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUGUUGAUAC-------------------UGGGCACGACCUCU ((((((((((....(((.....(((...((((......)))).))))))....((((((((.......))))))))..)))))))..-------------------..)))......... ( -30.70) >DroEre_CAF1 2073 101 - 1 GCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUGUUGAUAC-------------------UGGGCACGACCUCU ((((((((((....(((.....(((...((((......)))).))))))....((((((((.......))))))))..)))))))..-------------------..)))......... ( -30.70) >DroYak_CAF1 2224 101 - 1 GCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUGUUGAUAC-------------------UGGGCACGACCUCU ((((((((((....(((.....(((...((((......)))).))))))....((((((((.......))))))))..)))))))..-------------------..)))......... ( -30.70) >DroPer_CAF1 2572 117 - 1 GCCAUCAAUAAACGCUGACAGAGCCAA-UGGCAGAGCAGCUGAGGCCAG-AAGAGCUCAUGACAUUACCGUGAACUAAUGUUGAUACGGGGGCCACCGGGGGCCACCGGGCACGACCCC- ((((((((((...((((.....(((..-.)))....)))).........-...((.(((((.......))))).))..))))))).(((.((((......)))).))))))........- ( -36.80) >consensus GCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUGUUGAUAC___________________UGGGCACGACCUCU ...(((((((....(((.....(((...((((......)))).))))))....((((((((.......))))))))..)))))))......................(((.....))).. (-25.82 = -25.27 + -0.55)

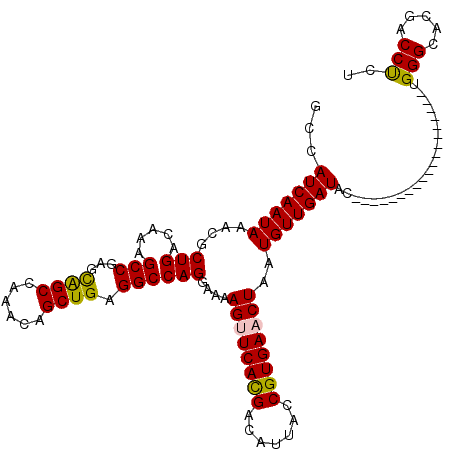
| Location | 1,206,456 – 1,206,569 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -24.55 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1206456 113 + 22407834 CAUUAGUUCACGGUAAUGUCGUGAACUUUUCCUGGCCUCAGCUGUUUGGCUGCUCGGCUUUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUU-UUCAAUUUAUUUUAUUUU--CCU---- ....(((((((((.....)))))))))......((.....((((...((((....))))....))))((..(((((.(((((....))))).-.)))))..))........--)).---- ( -26.60) >DroPse_CAF1 2603 118 + 1 CAUUAGUUCACGGUAAUGCCAUGAGCUCUU-CUGGCCUCAGCUGCUCUGCCA-UUGGCUCUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUUUUUCAAUUUAUUUUGUCUUUUUUGCCUU ...........((((((((((((((..(..-..)..))).(((((...(((.-..)))...).))))........)))))).(((((((...........)))))))......))))).. ( -28.00) >DroSim_CAF1 2160 113 + 1 CAUUAGUUCACGGUAAUGUCGUGAACUUUUCCUGGCCUCAGCUGUUUGGCUGCUCGGCUUUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUU-UUCAAUUUAUUUUAUUUU--CCG---- ....(((((((((.....)))))))))......((.....((((...((((....))))....))))((..(((((.(((((....))))).-.)))))..))........--)).---- ( -27.10) >DroEre_CAF1 2094 113 + 1 CAUUAGUUCACGGUAAUGUCGUGAACUUUUCCUGGCCUCAGCUGUUUGGCUGCUCGGCUUUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUU-UUCAAUUUAUUUUAUUUU--CCG---- ....(((((((((.....)))))))))......((.....((((...((((....))))....))))((..(((((.(((((....))))).-.)))))..))........--)).---- ( -27.10) >DroYak_CAF1 2245 113 + 1 CAUUAGUUCACGGUAAUGUCGUGAACUUUUCCUGGCCUCAGCUGUUUGGCUGCUCGGCUUUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUU-UUCAAUUUAUUUUAUUUU--CCG---- ....(((((((((.....)))))))))......((.....((((...((((....))))....))))((..(((((.(((((....))))).-.)))))..))........--)).---- ( -27.10) >DroPer_CAF1 2611 117 + 1 CAUUAGUUCACGGUAAUGUCAUGAGCUCUU-CUGGCCUCAGCUGCUCUGCCA-UUGGCUCUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUUUUUCAAUUUAUUUUGUCUUUUUUGCCU- ...........((((((((((((((..(..-..)..))).(((((...(((.-..)))...).))))........)))))).(((((((...........)))))))......))))).- ( -25.30) >consensus CAUUAGUUCACGGUAAUGUCGUGAACUUUUCCUGGCCUCAGCUGUUUGGCUGCUCGGCUUUGUCAGCGUUUAUUGAUGGCAUCAAAGUGUUU_UUCAAUUUAUUUUAUUUU__CCG____ ....(((((((((.....))))))))).............((((...((((....))))....))))((..(((((.(((((....)))))...)))))..))................. (-24.55 = -24.63 + 0.08)
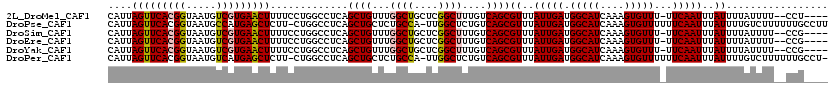
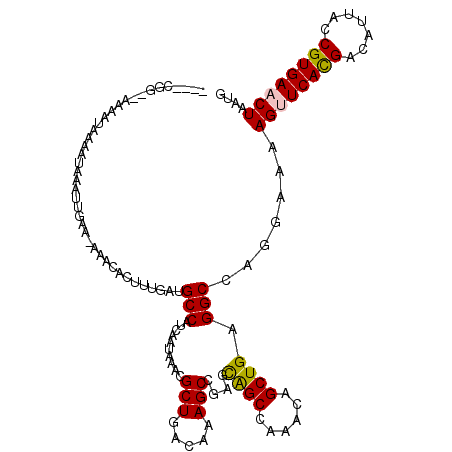
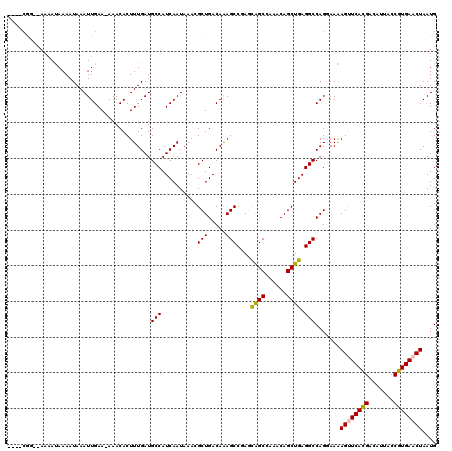
| Location | 1,206,456 – 1,206,569 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1206456 113 - 22407834 ----AGG--AAAAUAAAAUAAAUUGAA-AAACACUUUGAUGCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUG ----...--............((..((-......))..)).((.(((........)))....(((...((((......)))).)))..))...((((((((.......)))))))).... ( -22.90) >DroPse_CAF1 2603 118 - 1 AAGGCAAAAAAGACAAAAUAAAUUGAAAAAACACUUUGAUGCCAUCAAUAAACGCUGACAGAGCCAA-UGGCAGAGCAGCUGAGGCCAG-AAGAGCUCAUGGCAUUACCGUGAACUAAUG ...............................(((..(((((((((.................(((..-.))).((((..(((....)))-....)))))))))))))..)))........ ( -25.70) >DroSim_CAF1 2160 113 - 1 ----CGG--AAAAUAAAAUAAAUUGAA-AAACACUUUGAUGCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUG ----...--............((..((-......))..)).((.(((........)))....(((...((((......)))).)))..))...((((((((.......)))))))).... ( -22.90) >DroEre_CAF1 2094 113 - 1 ----CGG--AAAAUAAAAUAAAUUGAA-AAACACUUUGAUGCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUG ----...--............((..((-......))..)).((.(((........)))....(((...((((......)))).)))..))...((((((((.......)))))))).... ( -22.90) >DroYak_CAF1 2245 113 - 1 ----CGG--AAAAUAAAAUAAAUUGAA-AAACACUUUGAUGCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUG ----...--............((..((-......))..)).((.(((........)))....(((...((((......)))).)))..))...((((((((.......)))))))).... ( -22.90) >DroPer_CAF1 2611 117 - 1 -AGGCAAAAAAGACAAAAUAAAUUGAAAAAACACUUUGAUGCCAUCAAUAAACGCUGACAGAGCCAA-UGGCAGAGCAGCUGAGGCCAG-AAGAGCUCAUGACAUUACCGUGAACUAAUG -..((........(((......)))........((((...(((.(((......((((.....(((..-.)))....))))))))))..)-))).))(((((.......)))))....... ( -20.10) >consensus ____CGG__AAAAUAAAAUAAAUUGAA_AAACACUUUGAUGCCAUCAAUAAACGCUGACAAAGCCGAGCAGCCAAACAGCUGAGGCCAGGAAAAGUUCACGACAUUACCGUGAACUAAUG ........................................(((..........(((.....)))....((((......)))).))).......((((((((.......)))))))).... (-19.86 = -19.53 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:45 2006