| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,385,641 – 11,385,743 |
| Length | 102 |
| Max. P | 0.694276 |

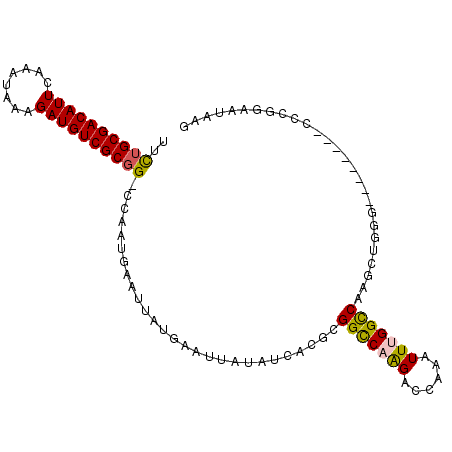
| Location | 11,385,641 – 11,385,743 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -19.14 |
| Energy contribution | -18.75 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11385641 102 + 22407834 CUUAUUCCGGG-CCUUAUUCCCAGCUUGGCCAAAUUUGGUCUUGGCCGCGUGAUAUAAUUCAUAAUUCAUUGG-CCGCGACAUCUUUAUUUGAAUGUCGCAGAA .........((-((.........((..((((((........))))))))((((......))))........))-))((((((((.......).))))))).... ( -31.10) >DroSim_CAF1 109613 102 + 1 CUUAUUCCGGG-CCUUAUUCCCAGCUUGGCCAAAUUUGGUCUUGGCCGCGUGAUAUAAUUCAUAAUUCAUUGG-CCGCGACAUCUUUAUUUGAAUGUCGCAGAA .........((-((.........((..((((((........))))))))((((......))))........))-))((((((((.......).))))))).... ( -31.10) >DroEre_CAF1 106239 95 + 1 CCUAUUCCGGG--------CCCAGCCUGGCCAAAUUUGGUCUUGGCCGCGUGAUAUAAUUCAUAAUUCAUUGG-CCGCGACAUCUUUAUUUGAAUGUCGCAGAA .........((--------((..(((.(((((....)))))..)))...((((......))))........))-))((((((((.......).))))))).... ( -30.90) >DroYak_CAF1 105963 103 + 1 CCUAUUCCCGGGUCCUAUUCCCAGCUUGGCCAAAUUUGGUCUUGGCCGCGUGAUAUAAUUCAUAAUUCAUUGG-CCGCGACAUCUUUAUUUGAAUGUCGCAGAA .........(((.......))).....((((((....(((....)))..((((......))))......))))-))((((((((.......).))))))).... ( -30.90) >DroAna_CAF1 91241 92 + 1 UUCUUUCCGGG-A-----------UCUGGCCAAAUUUGGUCUGGGCCGCGUGAUAUAAUUCAUAAUUCAUUCGCCCGCGACAUCUUUAUUUGAAUGUCGCAAAA ...........-.-----------...(((((....))))).((((.(.((((.............)))).)))))((((((((.......).))))))).... ( -25.02) >DroPer_CAF1 126557 87 + 1 UCUA----------------CGGAUUUGACCAAAUUUGGUCCGGACCGCGUGAUAUAAUUCAUAAUUCAUUCG-CAGCGACAUCUUUAUUUGAAUGUCGCAAAA ....----------------(((.(..(((((....)))))..).))).((((.................)))-).((((((((.......).))))))).... ( -22.43) >consensus CCUAUUCCGGG________CCCAGCUUGGCCAAAUUUGGUCUUGGCCGCGUGAUAUAAUUCAUAAUUCAUUGG_CCGCGACAUCUUUAUUUGAAUGUCGCAGAA .......................(((.(((((....)))))..)))...((((......)))).............((((((((.......).))))))).... (-19.14 = -18.75 + -0.39)

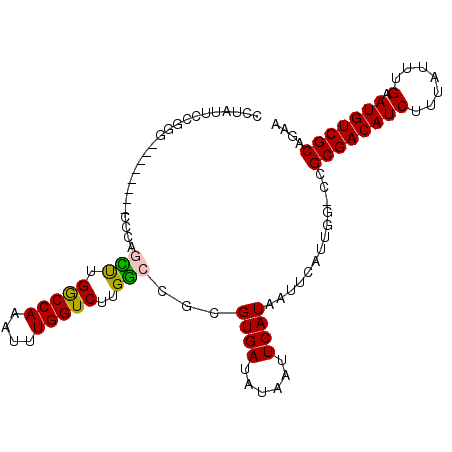
| Location | 11,385,641 – 11,385,743 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11385641 102 - 22407834 UUCUGCGACAUUCAAAUAAAGAUGUCGCGG-CCAAUGAAUUAUGAAUUAUAUCACGCGGCCAAGACCAAAUUUGGCCAAGCUGGGAAUAAGG-CCCGGAAUAAG ((((((((((((........))))))))((-((.........(((......))).(((((((((......)))))))..)).........))-)).)))).... ( -30.80) >DroSim_CAF1 109613 102 - 1 UUCUGCGACAUUCAAAUAAAGAUGUCGCGG-CCAAUGAAUUAUGAAUUAUAUCACGCGGCCAAGACCAAAUUUGGCCAAGCUGGGAAUAAGG-CCCGGAAUAAG ((((((((((((........))))))))((-((.........(((......))).(((((((((......)))))))..)).........))-)).)))).... ( -30.80) >DroEre_CAF1 106239 95 - 1 UUCUGCGACAUUCAAAUAAAGAUGUCGCGG-CCAAUGAAUUAUGAAUUAUAUCACGCGGCCAAGACCAAAUUUGGCCAGGCUGGG--------CCCGGAAUAGG ((((((((((((........))))))))((-((.........(((......)))..(((((..(.(((....))))..)))))))--------)).)))).... ( -30.70) >DroYak_CAF1 105963 103 - 1 UUCUGCGACAUUCAAAUAAAGAUGUCGCGG-CCAAUGAAUUAUGAAUUAUAUCACGCGGCCAAGACCAAAUUUGGCCAAGCUGGGAAUAGGACCCGGGAAUAGG ..((((((((((........))))))))))-((........................(((((((......)))))))...(((((.......))))).....)) ( -31.10) >DroAna_CAF1 91241 92 - 1 UUUUGCGACAUUCAAAUAAAGAUGUCGCGGGCGAAUGAAUUAUGAAUUAUAUCACGCGGCCCAGACCAAAUUUGGCCAGA-----------U-CCCGGAAAGAA ....((((((((........))))))))(((((..(((..((.....))..))))))((((.(((.....)))))))...-----------.-.))........ ( -23.00) >DroPer_CAF1 126557 87 - 1 UUUUGCGACAUUCAAAUAAAGAUGUCGCUG-CGAAUGAAUUAUGAAUUAUAUCACGCGGUCCGGACCAAAUUUGGUCAAAUCCG----------------UAGA ....((((((((........))))))))..-........................((((....(((((....)))))....)))----------------)... ( -23.60) >consensus UUCUGCGACAUUCAAAUAAAGAUGUCGCGG_CCAAUGAAUUAUGAAUUAUAUCACGCGGCCAAGACCAAAUUUGGCCAAGCUGGG________CCCGGAAUAAG ..((((((((((........))))))))))...........................(((((((......)))))))........................... (-19.99 = -19.85 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:49 2006