
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
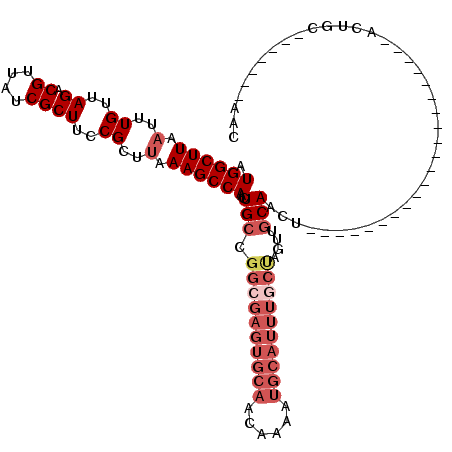
| Location | 11,378,045 – 11,378,182 |
| Length | 137 |
| Max. P | 0.921871 |

| Location | 11,378,045 – 11,378,143 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -18.39 |
| Energy contribution | -20.45 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11378045 98 + 22407834 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACUGC---------------UGCAACUGC-------AAC .((((((.(..((..((.((....))))..))..).))))))..(((.((((((((((......))))))))))(((((((.....---------------)))))))))-------).. ( -34.10) >DroSec_CAF1 95120 89 + 1 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCA------------------------ACUGC-------AAC .((((((.(..((..((.((....))))..))..).))))))......((((((((((......)))))))))).(((((.------------------------...))-------))) ( -28.90) >DroSim_CAF1 100045 89 + 1 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCA------------------------ACUGC-------AAC .((((((.(..((..((.((....))))..))..).))))))......((((((((((......)))))))))).(((((.------------------------...))-------))) ( -28.90) >DroEre_CAF1 98634 95 + 1 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACU------------------GCAACUGC-------AAC .((((((.(..((..((.((....))))..))..).))))))..(((.((((((((((......))))))))))((((((....------------------))))))))-------).. ( -33.30) >DroYak_CAF1 97879 76 + 1 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUG-------------CAAAAUGCAUUUGCUAGUUGCAACU------------------------GC-------AAC .((((((.(..((..((.((....))))..))..).))))))....-------------.....((((.((((.....)))).)------------------------))-------).. ( -20.00) >DroAna_CAF1 85070 120 + 1 AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUCAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUUCCAGUUGCAACAGCCCCAGCCAGAGGAACUGCAACUGCGGCAGGUAAC .((((((....((..((.((....))))..))....)))))).(((((((.(((((((......))))))).))(((((((....((.(.....).))...)))))))..)))))..... ( -40.10) >consensus AUGGCUUAAUUUGUUAGACGUUAUCGCUUCCGCUUAAAGCCAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACU_____________________ACUGC_______AAC .((((((.(..((..((.((....))))..))..).))))))..(((.((((((((((......))))))))))....)))....................................... (-18.39 = -20.45 + 2.06)


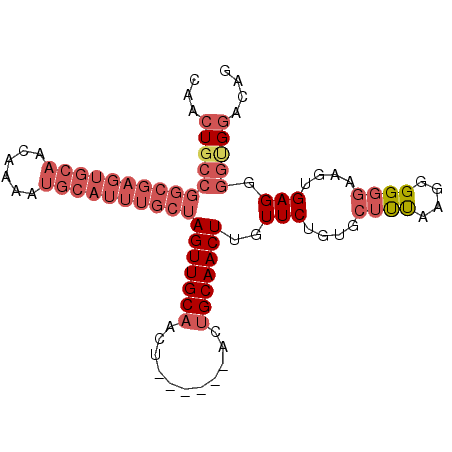
| Location | 11,378,085 – 11,378,182 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -17.28 |
| Energy contribution | -20.08 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11378085 97 + 22407834 CAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACUGCUGCAACUGCAACUUGUUCUGUGCUUUAAGGGGUGAAGUGAGGGGCGGACAG ...(((((((((((((((......))))))))))(((((((.....))))))).......(((((.(((((((.....))))))).))))))).))) ( -34.70) >DroSec_CAF1 95160 88 + 1 CAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCA---------ACUGCAACUUGUUCUGUGCUUUAAGGGGGGAAGUGAGGGGUGGACAG ...(..((((((((((((......))))))))))((((((.---------...))))))..((((.(.(.....).).))))......))..).... ( -29.30) >DroSim_CAF1 100085 88 + 1 CAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCA---------ACUGCAACUUGUUCUGUGCUUUAAGGGGGGAAGUGAGGGGUGGACAG ...(..((((((((((((......))))))))))((((((.---------...))))))..((((.(.(.....).).))))......))..).... ( -29.30) >DroEre_CAF1 98674 92 + 1 CAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACU---GCAACUGCAACUCGUUCUCUGCUCCAGCGGGG--AGUGAGGGGGGCACAG ....((((((((((((((......))))))))))((((((....---)))))).(..((((((((((((....))))))--))))))..)))))... ( -43.40) >DroYak_CAF1 97919 75 + 1 CAACUG-------------CAAAAUGCAUUUGCUAGUUGCAACU---------GCAACUUGUUCUGGUCUCCAAGGGGGGCGGUGAGGGGUGGACAG ((((((-------------((((.....))))).)))))...(.---------.(..(((...(((.((((....)))).))).)))..)..).... ( -23.20) >consensus CAACUGCCGGCGAGUGCAACAAAAUGCAUUUGCUAGUUGCAACU______ACUGCAACUUGUUCUGUGCUUUAAGGGGGGAAGUGAGGGGUGGACAG ...(((((((((((((((......))))))))))(((((((...........)))))))..(((....((((....))))....))).))))).... (-17.28 = -20.08 + 2.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:46 2006