
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,376,548 – 11,376,739 |
| Length | 191 |
| Max. P | 0.992824 |

| Location | 11,376,548 – 11,376,668 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -44.16 |
| Consensus MFE | -32.80 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11376548 120 + 22407834 CGUAACUCACUCCCCUCGCCUAACAAAUGGAGUUCGAGUUGGAGUUCUAGAUAGAGCGGAUGGGGAGGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGCGUGG .........(((((((((((((.....((((..((......))..))))..))).))))..))))))((((((((.(((.(.......).))).))))))))((((........)))).. ( -43.20) >DroSec_CAF1 93624 120 + 1 CGUAACUCACUCUCCUCACCCCGCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGUACAGAACGGAGUGG .......(((((.(((((..((((...((((..((......))..))))......)))).))))).(((((((((.(((.(.......).))).)))))))))(((.....)))))))). ( -42.90) >DroSim_CAF1 98533 120 + 1 CGUAACUCACUUCCCUCGCCCCUCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAACGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGG .......((((((..(((((((((.......((((...((((....))))...)))).)).)))))(((((((((.(((.(.......).))).)))))))))......))..)))))). ( -44.20) >DroEre_CAF1 97192 120 + 1 CGUAACUCACUCUCCUCGCCCCCCAAAUGGAGUUCGAGGUGGAGUUGCAGAUAGCGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGCAGUGG .(((((((.(...(((((..(.((....)).)..))))).)))))))).......(((..((..(((((((((((.(((.(.......).))).))))))))).)).))...)))..... ( -47.10) >DroYak_CAF1 96316 120 + 1 AGUAACUCACUCUCCUCGCCCCUCAAAUGGAGUUCGAGGUGGAGUUGCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGUGUGG .(((((((.(...(((((...(((.....)))..))))).))))))))...((.(.((..((..(((((((((((.(((.(.......).))).))))))))).)).))...)).).)). ( -43.40) >consensus CGUAACUCACUCUCCUCGCCCCUCAAAUGGAGUUCGAGUUGGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGG .......(((((((((((((((......)).((((...((((....))))...)))))).)))))..((((((((.(((.(.......).))).))))))))...........)))))). (-32.80 = -33.60 + 0.80)

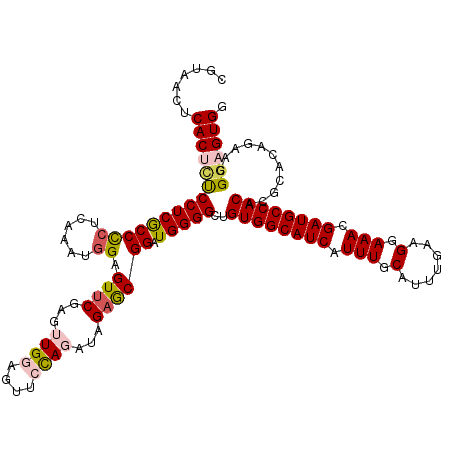

| Location | 11,376,548 – 11,376,668 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -29.66 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11376548 120 - 22407834 CCACGCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACCUCCCCAUCCGCUCUAUCUAGAACUCCAACUCGAACUCCAUUUGUUAGGCGAGGGGAGUGAGUUACG ..((((........))))((((((((((((.(.......).))))))))))))((((((...(((.(((.(..((..((......))..)).....).)))))).))))))......... ( -35.80) >DroSec_CAF1 93624 120 - 1 CCACUCCGUUCUGUACGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCCAACUCGAACUCCAUUUGCGGGGUGAGGAGAGUGAGUUACG .((((((((.....)))(((((((((((((.(.......).))))))))))))).((.(((((((......((((..((......))..))))...)).))))).)).)))))....... ( -39.60) >DroSim_CAF1 98533 120 - 1 CCACUCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGUUCUAUCUGGAACUCCAACUCGAACUCCAUUUGAGGGGCGAGGGAAGUGAGUUACG .((((((.(((......(((((((((((((.(.......).)))))))))))))(((((....(((((....)))))......(((((.....))))))))))))))).))))....... ( -44.10) >DroEre_CAF1 97192 120 - 1 CCACUGCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCGCUAUCUGCAACUCCACCUCGAACUCCAUUUGGGGGGCGAGGAGAGUGAGUUACG ....((((....((((((((((((((((((.(.......).)))))))))))))........)))))....))))((((..(((((..((((....))))..))))).))))........ ( -48.10) >DroYak_CAF1 96316 120 - 1 CCACACCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGCAACUCCACCUCGAACUCCAUUUGAGGGGCGAGGAGAGUGAGUUACU ...(((..((((.....(((((((((((((.(.......).)))))))))))))(((((....((.......)).........(((((.....)))))))))).))))..)))....... ( -39.30) >consensus CCACUCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCCAACUCGAACUCCAUUUGAGGGGCGAGGAGAGUGAGUUACG ......(((.....)))(((((((((((((.(.......).)))))))))))))....................((((((..((((..((((......))))))))....).)))))... (-29.66 = -29.94 + 0.28)

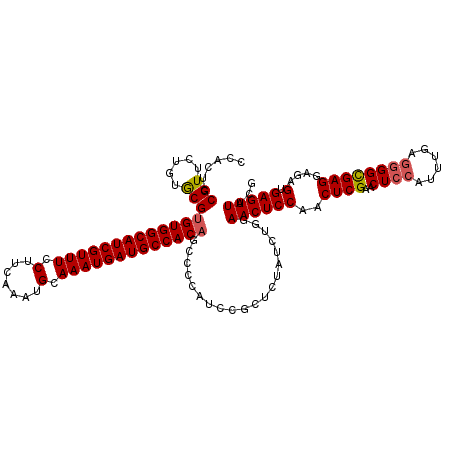
| Location | 11,376,588 – 11,376,699 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
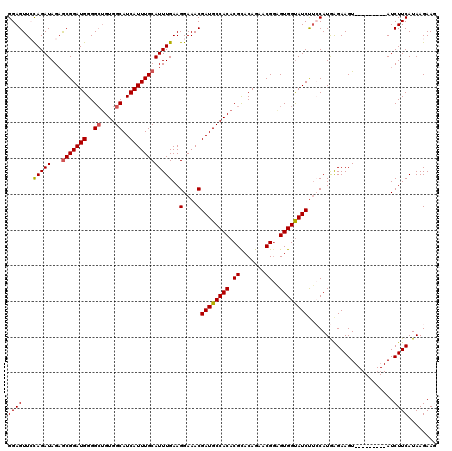
>2L_DroMel_CAF1 11376588 111 + 22407834 GGAGUUCUAGAUAGAGCGGAUGGGGAGGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGCGUGGUAUCCGCCAUGAGAAGU---------AUCUUCAUAAGAAG ...(((((....)))))..(((((((.((((((((.(((.(.......).))).))))))))((((........))))....))).))))..((((.---------..))))........ ( -34.20) >DroSec_CAF1 93664 111 + 1 GGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGUACAGAACGGAGUGGUAUCUUCCAUGAGAAGU---------AUCUUCAUAAGAAG ((((((((...(((((((((((..((....))..))))))).))))..))))..((((((((.(((.....)))..))))))))))))....((((.---------..))))........ ( -35.30) >DroSim_CAF1 98573 111 + 1 GGAGUUCCAGAUAGAACGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGGUAUCUUCCAUGAGAAGU---------AUCUUCAUAAGAAG ((....))........((..((..(((((((((((.(((.(.......).))).))))))))).)).))...))(((.((((.((((.....)))))---------))))))........ ( -30.70) >DroEre_CAF1 97232 110 + 1 GGAGUUGCAGAUAGCGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGCAGUGGUAUCUU-CAUAAGAAGU---------AUCUUCAUAAGAAA ((((.(((.......(((((((..((....))..)))))))...(((((....)((((((((..((.......)).))))))))))-))......))---------).))))........ ( -34.50) >DroYak_CAF1 96356 119 + 1 GGAGUUGCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGUGUGGCAUCUU-CAUAGGAAGUAUGUUCAGUAUCUUCAUAAGAAA ((((.(((.......(((((((..((....))..)))))))...(((((....)((((((((((..........))))))))))..-.............))))))).))))........ ( -39.30) >consensus GGAGUUCCAGAUAGAGCGGAUGGGGCUGUGGCAUCAUUUGCAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGGUAUCUUCCAUGAGAAGU_________AUCUUCAUAAGAAG ((((...(((((...(((((((..((....))..))))))))))))..(....)((((((((.((.......))..))))))))........................))))........ (-26.96 = -27.04 + 0.08)

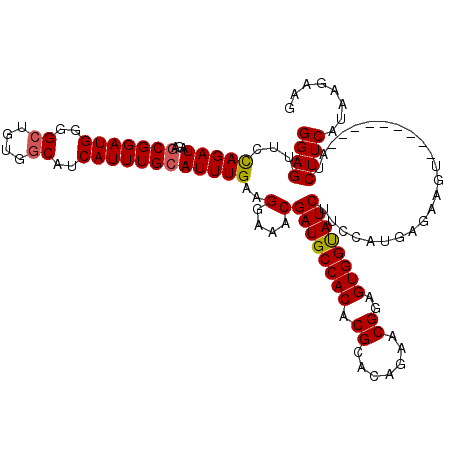
| Location | 11,376,588 – 11,376,699 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -24.44 |
| Energy contribution | -23.92 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
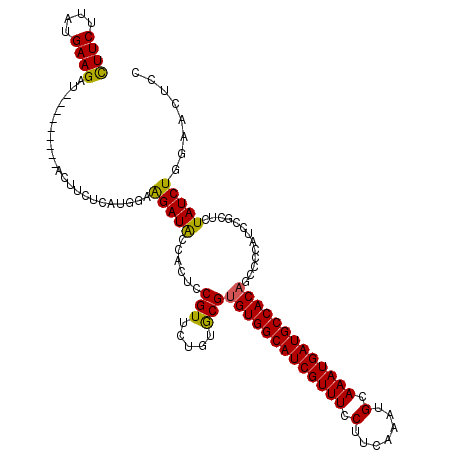
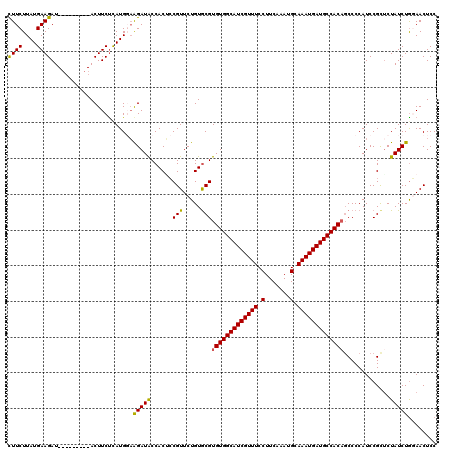
>2L_DroMel_CAF1 11376588 111 - 22407834 CUUCUUAUGAAGAU---------ACUUCUCAUGGCGGAUACCACGCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACCUCCCCAUCCGCUCUAUCUAGAACUCC ((((....))))..---------.((......(((((((...((((........))))((((((((((((.(.......).))))))))))))......)))))))......))...... ( -34.50) >DroSec_CAF1 93664 111 - 1 CUUCUUAUGAAGAU---------ACUUCUCAUGGAAGAUACCACUCCGUUCUGUACGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCC (((((.((((.((.---------...)))))))))))..........(((((.....(((((((((((((.(.......).)))))))))))))((.......)).......)))))... ( -31.10) >DroSim_CAF1 98573 111 - 1 CUUCUUAUGAAGAU---------ACUUCUCAUGGAAGAUACCACUCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGUUCUAUCUGGAACUCC (((((.((((.((.---------...))))))))))).................((.(((((((((((((.(.......).))))))))))))))).......(((((....)))))... ( -32.40) >DroEre_CAF1 97232 110 - 1 UUUCUUAUGAAGAU---------ACUUCUUAUG-AAGAUACCACUGCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCGCUAUCUGCAACUCC ..((((...((((.---------...))))...-))))......((((....((((((((((((((((((.(.......).)))))))))))))........)))))....))))..... ( -33.20) >DroYak_CAF1 96356 119 - 1 UUUCUUAUGAAGAUACUGAACAUACUUCCUAUG-AAGAUGCCACACCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGCAACUCC .......(((((..((....((((.....))))-..((((((((((.((.....))))))))))))))...)))))..((((.(((..((.............))..))).))))..... ( -29.82) >consensus CUUCUUAUGAAGAU_________ACUUCUCAUGGAAGAUACCACUCCGUUCUGUGCGUGUGGCAUCGUUUCCUUCAAAUGCAAAUGAUGCCACAGCCCCAUCCGCUCUAUCUGGAACUCC ((((....)))).......................(((((......(((.....)))(((((((((((((.(.......).))))))))))))).............)))))........ (-24.44 = -23.92 + -0.52)

| Location | 11,376,628 – 11,376,739 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
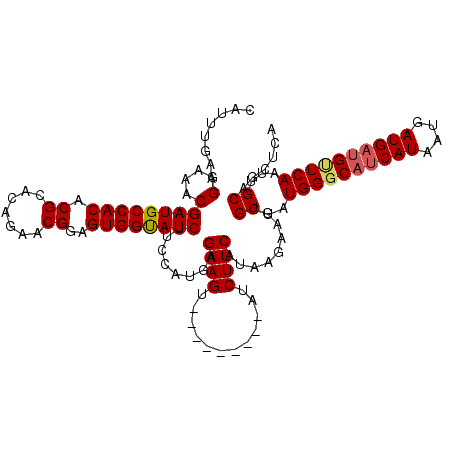
>2L_DroMel_CAF1 11376628 111 + 22407834 CAUUUGAAGGAAACGAUGCCACACGCACAGAACGGCGUGGUAUCCGCCAUGAGAAGU---------AUCUUCAUAAGAAGCGUUAUGGGCAUCAUAAUGAUGAUGUUCAAUUGCAGCUCA ........((....((((((((.((.......))..))))))))..)).((((..((---------(.((((....)))).....((((((((((....))))))))))..)))..)))) ( -32.90) >DroSec_CAF1 93704 111 + 1 CAUUUGAAGGAAACGAUGCCACACGUACAGAACGGAGUGGUAUCUUCCAUGAGAAGU---------AUCUUCAUAAGAAGCGUUAUGGGCAUCAUAAUGAUGAUGCUCAAUUGCAGCUCA ........((((..((((((((.(((.....)))..)))))))))))).((((..((---------(.((((....)))).....((((((((((....))))))))))..)))..)))) ( -39.00) >DroSim_CAF1 98613 111 + 1 CAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGGUAUCUUCCAUGAGAAGU---------AUCUUCAUAAGAAGCGUUAUGGGCAUCAUAAUGAUGAUGUUCAAUUGCAGCUCA ........((((..((((((((.((.......))..)))))))))))).((((..((---------(.((((....)))).....((((((((((....))))))))))..)))..)))) ( -34.60) >DroEre_CAF1 97272 110 + 1 CAUUUGAAGGAAACGAUGCCACACGCACAGAACGCAGUGGUAUCUU-CAUAAGAAGU---------AUCUUCAUAAGAAACGUUAUGGGCUUCAUAAUGAUGAUGUUCAAUUGCAGCUCA ....((((((....((((((((..((.......)).))))))))((-(....)))..---------.))))))............((((((.((...(((......)))..)).)))))) ( -27.00) >DroYak_CAF1 96396 119 + 1 CAUUUGAAGGAAACGAUGCCACACGCACAGAACGGUGUGGCAUCUU-CAUAGGAAGUAUGUUCAGUAUCUUCAUAAGAAACGUUAUGGCCACCAUAAUGAUGAUGUUCAAUUGCAGCUCA ((.((((((....)((((((((((..........)))))))))).(-(((..(((((((.....))).))))........((((((((...))))))))))))..))))).))....... ( -35.00) >consensus CAUUUGAAGGAAACGAUGCCACACGCACAGAACGGAGUGGUAUCUUCCAUGAGAAGU_________AUCUUCAUAAGAAGCGUUAUGGGCAUCAUAAUGAUGAUGUUCAAUUGCAGCUCA ........(....)((((((((.((.......))..))))))))........((((............)))).........((..((((((((((....))))))))))...))...... (-26.42 = -26.74 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:44 2006