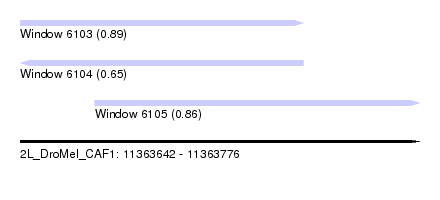
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,363,642 – 11,363,776 |
| Length | 134 |
| Max. P | 0.893128 |

| Location | 11,363,642 – 11,363,737 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -10.70 |
| Energy contribution | -13.03 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11363642 95 + 22407834 UUCCCACUUUU---------UUUUUCGCUGCCUC----CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAA ...........---------..........((((----((..((((..((..((((((.....................))))))...))..))))))))))...---... ( -21.90) >DroSec_CAF1 80904 95 + 1 UUCCCACUUUU---------UUUUUCGCUGCCUC----CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAA ...........---------..........((((----((..((((..((..((((((.....................))))))...))..))))))))))...---... ( -21.90) >DroSim_CAF1 85615 95 + 1 UUCCCACUUUU---------UUUUUCGCUGCCUC----CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAA ...........---------..........((((----((..((((..((..((((((.....................))))))...))..))))))))))...---... ( -21.90) >DroEre_CAF1 84430 92 + 1 UUCCCACUUU------------UUUCGCUACCUC----CGCAGCCGUUUUUAUUUCCCAUAUUUUCCUUGGCUUUUUUCGGGAAAGUCAUUUCGGUGGGAGGAAA---AAA ..........------------........((((----(...((((......((((((.....................)))))).......)))).)))))...---... ( -21.42) >DroWil_CAF1 88041 98 + 1 UC-CUACAUUUCGGUUUCUUUCUUUCAUUUUCUUCUAUUUGCACCCUUUCCUUCUCCCAGGGUUUCUUA-UCUCUUGUUGGCAUAUUUAU-------GGAUC-AA---AAA ..-........................((((..(((((.((((((((...........)))))......-...(.....)))).....))-------)))..-))---)). ( -8.90) >DroYak_CAF1 82644 98 + 1 UUCCCACUUUU-----------UUUCGCUGCCUC-AACCUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUU-UUUCGGGAAAGUCAAUGCGGUGGGAGGAAAAAAAAA .......((((-----------((((.((.((..-.......(((((.....((((((................-....))))))......))))))).)))))))))).. ( -21.55) >consensus UUCCCACUUUU_________UUUUUCGCUGCCUC____CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA___AAA ....................................................((((((((.......(((((((((.....)))))))))....))))))))......... (-10.70 = -13.03 + 2.33)

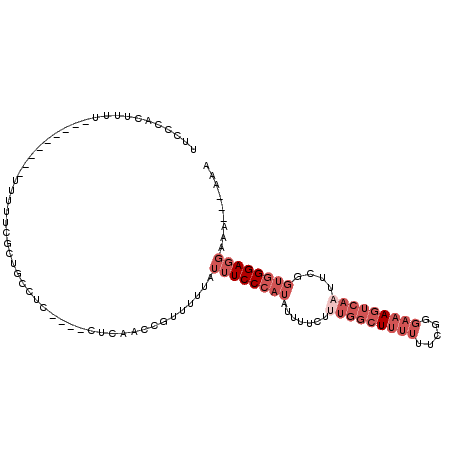
| Location | 11,363,642 – 11,363,737 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -12.11 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11363642 95 - 22407834 UUU---UUUCCUCCCACCGAAUUGACUUUCCCGAAAAAAGCCAAAGAAAAUAUGGGAAAUAAAAACGGUUGAG----GAGGCAGCGAAAAA---------AAAAGUGGGAA (((---(((((((((((((..((...(((((((...................)))))))..))..))).)).)----))))....))))))---------........... ( -18.91) >DroSec_CAF1 80904 95 - 1 UUU---UUUCCUCCCACCGAAUUGACUUUCCCGAAAAAAGCCAAAGAAAAUAUGGGAAAUAAAAACGGUUGAG----GAGGCAGCGAAAAA---------AAAAGUGGGAA (((---(((((((((((((..((...(((((((...................)))))))..))..))).)).)----))))....))))))---------........... ( -18.91) >DroSim_CAF1 85615 95 - 1 UUU---UUUCCUCCCACCGAAUUGACUUUCCCGAAAAAAGCCAAAGAAAAUAUGGGAAAUAAAAACGGUUGAG----GAGGCAGCGAAAAA---------AAAAGUGGGAA (((---(((((((((((((..((...(((((((...................)))))))..))..))).)).)----))))....))))))---------........... ( -18.91) >DroEre_CAF1 84430 92 - 1 UUU---UUUCCUCCCACCGAAAUGACUUUCCCGAAAAAAGCCAAGGAAAAUAUGGGAAAUAAAAACGGCUGCG----GAGGUAGCGAAA------------AAAGUGGGAA ...---.....((((((.........(((((((.......(....)......)))))))........(((((.----...)))))....------------...)))))). ( -23.62) >DroWil_CAF1 88041 98 - 1 UUU---UU-GAUCC-------AUAAAUAUGCCAACAAGAGA-UAAGAAACCCUGGGAGAAGGAAAGGGUGCAAAUAGAAGAAAAUGAAAGAAAGAAACCGAAAUGUAG-GA (((---((-(...(-------((....)))....)))))).-......(((((...........)))))............................((........)-). ( -8.90) >DroYak_CAF1 82644 98 - 1 UUUUUUUUUCCUCCCACCGCAUUGACUUUCCCGAAA-AAGCCAAAGAAAAUAUGGGAAAUAAAAACGGUUGAGGUU-GAGGCAGCGAAA-----------AAAAGUGGGAA .((((((((((((..((((..((...(((((((...-...(....)......)))))))..))..)))).)))(((-(...))))))))-----------)))))...... ( -19.10) >consensus UUU___UUUCCUCCCACCGAAUUGACUUUCCCGAAAAAAGCCAAAGAAAAUAUGGGAAAUAAAAACGGUUGAG____GAGGCAGCGAAAAA_________AAAAGUGGGAA ...........((((((.........(((((((...................)))))))........((((..........))))...................)))))). (-12.11 = -12.64 + 0.53)


| Location | 11,363,667 – 11,363,776 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -26.84 |
| Energy contribution | -28.08 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11363667 109 + 22407834 CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAAGCGUGGGGCUGAGGUUCACCUUUGCCG-GAGUUUUUCCAU (((..((((((((.((((((((.......(((((((((.....)))))))))....))))))))..)---)))))).).(((.((((....)))).))).-)))......... ( -35.20) >DroSec_CAF1 80929 109 + 1 CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAAGCGUGGGGCUGAGGUUCACCUUUGCCG-GAGUUUUUCCAU (((..((((((((.((((((((.......(((((((((.....)))))))))....))))))))..)---)))))).).(((.((((....)))).))).-)))......... ( -35.20) >DroSim_CAF1 85640 109 + 1 CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA---AAAGCGUGGGGCUGAGGUUCACCUUUGCCG-GAGUUUUUCCAU (((..((((((((.((((((((.......(((((((((.....)))))))))....))))))))..)---)))))).).(((.((((....)))).))).-)))......... ( -35.20) >DroEre_CAF1 84452 109 + 1 CGCAGCCGUUUUUAUUUCCCAUAUUUUCCUUGGCUUUUUUCGGGAAAGUCAUUUCGGUGGGAGGAAA---AAAGCGUGGGGCUCGGGCUCACCUUCGGCG-GAGUUUUUCCAU ....(((((((((.((((((((........((((((((.....)))))))).....))))))))..)---)))))(((((.(...).)))))....)))(-(((...)))).. ( -33.42) >DroYak_CAF1 82670 112 + 1 CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUU-UUUCGGGAAAGUCAAUGCGGUGGGAGGAAAAAAAAAGCGUGGGGCUGAGGUUCACCUUUUGCGGGAGUUUUUCCAU ................((((((.......(((((((-((....)))))))))....))))))(((((((....(((..(((.((.....)))))..))).....))))))).. ( -32.80) >consensus CUCAACCGUUUUUAUUUCCCAUAUUUUCUUUGGCUUUUUUCGGGAAAGUCAAUUCGGUGGGAGGAAA___AAAGCGUGGGGCUGAGGUUCACCUUUGCCG_GAGUUUUUCCAU (((..(((((((..((((((((.......(((((((((.....)))))))))....))))))))......)))))).).(((.((((....)))).)))..)))......... (-26.84 = -28.08 + 1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:36 2006