
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,339,986 – 11,340,087 |
| Length | 101 |
| Max. P | 0.742384 |

| Location | 11,339,986 – 11,340,087 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.37 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11339986 101 - 22407834 -CCAA----CUG---GUGGCUGGCCAUAAAAUCAACACUUUGGUUAGCAA-AACAAACCGUUUAAUGCAAAA-CAAACAAGAGGCGACAACAACUAGCC------ACACCUGGCCAA-- -(((.----..(---(((((((((((..............))))))))..-........((((......)))-)........(((...........)))------.)))))))....-- ( -22.64) >DroVir_CAF1 53037 92 - 1 ----------CA---GCGGUCGGCCAUAAAAUCAACACUUUGGUUAGCAA-AACAAACCGUUUAAUGCAAAA-CAAACA---GUCAACAAC---UAGAU------ACACCUGGUCAGCA ----------..---((.((.(((.....((((((....)))))).((((-(((.....))))..)))....-......---))).)).((---(((..------....)))))..)). ( -14.60) >DroPse_CAF1 71983 115 - 1 UCGGCAUCGUCG---GUGGUCGGCCAUAAAAUCAACACUUUGGUUAGCAA-AACAAACCGUUUAAUGCAAAAGCAAACAAGAGUCGACAACAACUAGCCGGCCACACACCUGGCCAACG ..(((.......---(((((((((............(((((((((.(...-..).))))(((...(((....)))))).)))))............))))))))).......))).... ( -31.49) >DroGri_CAF1 68260 96 - 1 ----------CG---GCGGUUGGCCAUAAAAUCAACACUUUGGUUAGCAAAAACAAACCGUUUAAUGCAAAA-CAAACA---GUCAACAACGGCCAGCU------ACACCUGGCCAACA ----------..---(..((((((.....((((((....)))))).(((.((((.....))))..)))....-......---))))))..)((((((..------....)))))).... ( -24.80) >DroAna_CAF1 51240 105 - 1 -CGGC----GUGGCGGCGGUCGGCCAUAAAAUCAACACUUUGGUUAGCAA-AACAAACCGUUUAAUGCAAAA-CAAACAGGAGGCGACAGCAACUAGCC------ACACCUGGCAAAA- -.((.----(((((.((.((((.((....((((((....)))))).((((-(((.....))))..)))....-.........)))))).)).....)))------)).))........- ( -30.00) >DroPer_CAF1 72450 114 - 1 -CGGCAUCGUCG---GUGGUCGGCCAUAAAAUCAACACUUUGGUUAGCAA-AACAAACCGUUUAAUGCAAAAGCAAACAAGAGUCGACAACAACUAGCCGGCCACACACCUGGCCAACG -.(((.......---(((((((((............(((((((((.(...-..).))))(((...(((....)))))).)))))............))))))))).......))).... ( -31.49) >consensus _CGGC_____CG___GCGGUCGGCCAUAAAAUCAACACUUUGGUUAGCAA_AACAAACCGUUUAAUGCAAAA_CAAACAAGAGUCGACAACAACUAGCC______ACACCUGGCCAAC_ .....................(((((...((((((....)))))).(((..(((.....)))...)))..........................................))))).... (-13.34 = -13.37 + 0.03)

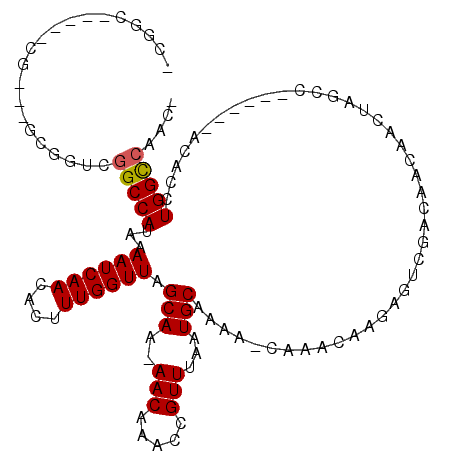

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:23 2006