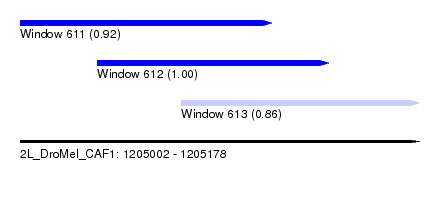
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,205,002 – 1,205,178 |
| Length | 176 |
| Max. P | 0.999806 |

| Location | 1,205,002 – 1,205,113 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.24 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
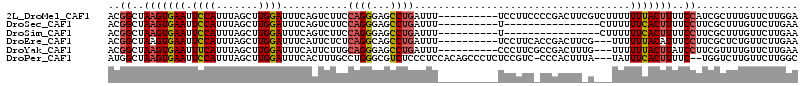
>2L_DroMel_CAF1 1205002 111 + 22407834 UCAGGCUCA------GAUUCAGAUUCCGAGCCUUACUCAUCCCCGGCACAUCAACAUCAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUC ..((((((.------((.......)).)))))).(((.((((..((((.........................))))..(((((((((.....))))))))).))))...))).... ( -26.61) >DroSim_CAF1 721 108 + 1 UCAGGCUCG------GAUUCAGAUUCCGAGCCUUACUCAUCCCCGGCACAUCAC---CAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUC ..(((((((------((.......))))))))).(((.((((..((((......---................))))..(((((((((.....))))))))).))))...))).... ( -32.85) >DroEre_CAF1 672 113 + 1 -CGGAUUCAGAUUCAGAUACAGAUUCCGAGCCUUGCUCAUCCCCUACACAUCCUCAGCAU---CACCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAUUCUCU -.((((...((.((.......)).)).((((...))))...........)))).((((..---........))))(((.(((((((((.....))))))))))))............ ( -24.40) >consensus UCAGGCUCA______GAUUCAGAUUCCGAGCCUUACUCAUCCCCGGCACAUCAACA_CAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUC ..((((((...................))))))..........................................(((.(((((((((.....))))))))))))............ (-18.35 = -18.24 + -0.11)

| Location | 1,205,036 – 1,205,138 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.73 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.51 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1205036 102 + 22407834 CCCCGGCACAUCAACAUCAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU----------UC-----CUUCCCCG ...........................(((((((.((..(((((((((.....)))))))))((((.........)))))).)).)))))..----------..-----........ ( -23.60) >DroPse_CAF1 741 106 + 1 ----------CCUCCAUCGUCGUCAUCAUCAUCUGCCAUGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCACUUUGCCUGGCCGUCUCCCUCCACAGCCCUCUCCAGCCCGUC-CCC ----------........((.((.........(.((((.(((.(((((((.((((.......)))).))))))).))))))).).........)).))...............-... ( -26.27) >DroSim_CAF1 755 90 + 1 CCCCGGCACAUCAC---CAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU----------U-------------- ....((.......)---).........(((((((.((..(((((((((.....)))))))))((((.........)))))).)).)))))..----------.-------------- ( -24.10) >DroEre_CAF1 711 99 + 1 CCCCUACACAUCCUCAGCAU---CACCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAUUCUCUCAGGCAGCCUGAUUU----------UC-----CUUCACCG ....................---....((((((((((..(((((((((.....))))))))).(((......)))....))))).)))))..----------..-----........ ( -25.10) >DroPer_CAF1 735 101 + 1 ----------CCUCCAUCGACGUCAUCAUCAUCUGCCAUGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCACUUUGCCUGGGCGUCUCCCUCCACAGCCCUCUC-----CGUC-CCC ----------........((((.................(((.(((((((.((((.......)))).))))))).))).((((.............))))....-----))))-... ( -30.22) >consensus CCCC__CACAUCACCAUCAUCAUCAUCAUCAGCUGCCACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCACUCUGCCAGGGAGCCUGAUUU__________UC_____CGUC_CCC ...................................(((.(((((((((.....)))))))))))).................................................... (-13.30 = -13.30 + -0.00)


| Location | 1,205,073 – 1,205,178 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -11.88 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
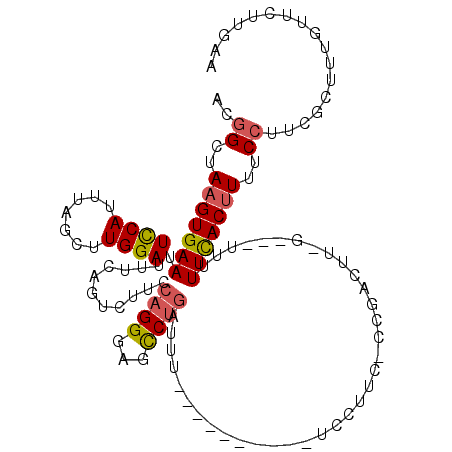
>2L_DroMel_CAF1 1205073 105 + 22407834 ACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU----------UCCUUCCCCGACUUCGUCUUUUUUUACUUUUCCAUCGCUUUGUUCUUGGA ..(((((((((.....))))))))).(((..((((.(((.....))).))))...----------)))......(((...)))............((((............)))) ( -22.30) >DroSec_CAF1 799 89 + 1 ACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU----------U----------------CUUUUUUCACUUUUCCUUCGCUUUGUUCUUGAA ..((..(((((((.((((.......))))..((((.(((.....))).))))...----------.----------------.....)))))))..))................. ( -19.30) >DroSim_CAF1 789 89 + 1 ACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU----------U----------------CUUUUUUCACUUUUCCUUCGCUUUGUUCUUGAA ..((..(((((((.((((.......))))..((((.(((.....))).))))...----------.----------------.....)))))))..))................. ( -19.30) >DroEre_CAF1 745 102 + 1 ACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAUUCUCUCAGGCAGCCUGAUUU----------UCCUUCACCGACUUCG---UUUUUUACAUUUCCUUCGCUCUGUUCUUGAA .(((...((((((.((((.......)))).))))))...(((((...)))))...----------.......)))......---............................... ( -17.10) >DroYak_CAF1 811 102 + 1 ACGGCUAAGUGAAUUUCAUUUAGCUUGGAUUUCAUUCUUGCAGGGAGCCUGAUUU----------CCCUUCGCCGACUUUG---UUUUUUACUUAUCCUUCGUUUUGUUCUUGAA .((((..((((((.((((.......)))).))))))...(.((((((......))----------)))).)))))......---............................... ( -20.90) >DroPer_CAF1 762 109 + 1 AUGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCACUUUGCCUGGGCGUCUCCCUCCACAGCCCUCUCCGUC-CCCACUUUA---UAUUUCACUUUUC--UGGUCUUGUUCUUGGC ..(((.(((((((.((((.......)))).))))))).))).((((.............))))........-.(((....(---((..(((......--)))...)))...))). ( -27.02) >consensus ACGGCUAAGUGAAUUCCAUUUAGCUUGGAUUUCAGUCUUCCAGGGAGCCUGAUUU__________UCCUUC_CCGACUU_G___UUUUUCACUUUUCCUUCGCUUUGUUCUUGAA ..((..(((((((.((((.......))))...........((((...))))....................................)))))))..))................. (-11.88 = -12.02 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:40 2006