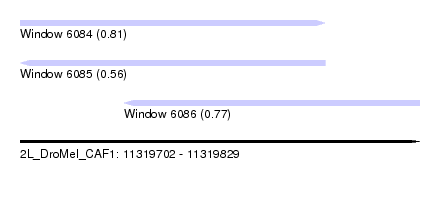
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,319,702 – 11,319,829 |
| Length | 127 |
| Max. P | 0.809884 |

| Location | 11,319,702 – 11,319,799 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -23.51 |
| Energy contribution | -25.60 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11319702 97 + 22407834 UGGCUCAGUGGUUGCGGCUGG-CGCAGUUUCA-----GCUUU--UUGCGCACCGCUGG--GGCACAGCGGUGAUUAAUUAAACCGACUGC---AACUCCAACUCAUGAUU .......(..(((((((.(((-(((((.....-----.....--)))))((((((((.--....))))))))..........))).))))---)))..)........... ( -34.90) >DroSec_CAF1 37094 97 + 1 UGGCUCAGUGGUUGCGGCUGG-CGCAGUUUUA-----GCUUU--UUGCGCAUCGCUGG--GGCACAGCGGUGAUUAAUUAAACCGACUGC---AGCUCCAACUCAUGAUU ....(((..(((((.((((..-.((((((...-----((...--..)).((((((((.--....))))))))............))))))---)))).)))))..))).. ( -33.80) >DroEre_CAF1 39045 92 + 1 -----CAGUGGUUGCGGCUGG-CGAAGUUUUA-----GCUUU--UUGCGCACCGCUGG--GACACAGCGGUGAUUAAUUAAACCGAGCGC---AGCUCCAGCUCAUGAUU -----..(((((((.((((((-(...((((..-----((...--..)).((((((((.--....)))))))).......))))...)).)---)))).)))).))).... ( -35.90) >DroYak_CAF1 37464 93 + 1 -----CAGUGGUUGCGGCUGGUCGCAGUUUUA-----GCUUU--UUGCGCACCGCUGG--GGCACAGCGGUGAUUAAUUAAAUCGAGCGC---AGCUCCAACUCAUGAUU -----..(((((((.(((((..(((.......-----((...--..)).((((((((.--....))))))))..............))))---)))).)))).))).... ( -35.00) >DroAna_CAF1 33273 96 + 1 ----UCAGUGUUUCUGUUCGCCGGCAGUUUGA-----GCUUUUUUUGUGCACCGCUGG--GGCACAGCGGUGAUUAAUUAAACCGACUGC---AGCUUCAACUCAUGAUU ----.(((.....)))...((..((((((.(.-----(.(((..(((..((((((((.--....))))))))..)))..)))))))))))---.)).............. ( -26.50) >DroPer_CAF1 50513 96 + 1 -----CAUU------GGCAGU-UGCAGUUGCAUUGGUGCUUU--UUGUGCACCGCUGUGUGCCCCAGCGGUGAUUAAUUAAACCGACUGCCGUGGCUCCAACUCAUGAUU -----..((------((.(((-..(....((((((((.....--(((..((((((((.......))))))))..)))....))))).))).)..)))))))......... ( -32.00) >consensus _____CAGUGGUUGCGGCUGG_CGCAGUUUUA_____GCUUU__UUGCGCACCGCUGG__GGCACAGCGGUGAUUAAUUAAACCGACUGC___AGCUCCAACUCAUGAUU .....((..(((((.((((....((((((........((.......)).((((((((.......))))))))............))))))...)))).)))))..))... (-23.51 = -25.60 + 2.09)


| Location | 11,319,702 – 11,319,799 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11319702 97 - 22407834 AAUCAUGAGUUGGAGUU---GCAGUCGGUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCGCAA--AAAGC-----UGAAACUGCG-CCAGCCGCAACCACUGAGCCA ..(((((.((((..(((---(((((((((((.......((((((((....--.)))))))).....--.))))-----)))..)))))-...))..)))))).))).... ( -31.22) >DroSec_CAF1 37094 97 - 1 AAUCAUGAGUUGGAGCU---GCAGUCGGUUUAAUUAAUCACCGCUGUGCC--CCAGCGAUGCGCAA--AAAGC-----UAAAACUGCG-CCAGCCGCAACCACUGAGCCA ..(((((.(((((.((.---((((.((((..........)))))))))).--)))))..((((...--...((-----.......)).-.....))))..)).))).... ( -26.60) >DroEre_CAF1 39045 92 - 1 AAUCAUGAGCUGGAGCU---GCGCUCGGUUUAAUUAAUCACCGCUGUGUC--CCAGCGGUGCGCAA--AAAGC-----UAAAACUUCG-CCAGCCGCAACCACUG----- ......(.(((((((((---((((..((((.....))))(((((((....--.)))))))))))..--..)))-----).........-))))))..........----- ( -28.50) >DroYak_CAF1 37464 93 - 1 AAUCAUGAGUUGGAGCU---GCGCUCGAUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCGCAA--AAAGC-----UAAAACUGCGACCAGCCGCAACCACUG----- .......((((..((((---((((..((((.....))))(((((((....--.)))))))))))..--..)))-----)..))))(((......)))........----- ( -31.00) >DroAna_CAF1 33273 96 - 1 AAUCAUGAGUUGAAGCU---GCAGUCGGUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCACAAAAAAAGC-----UCAAACUGCCGGCGAACAGAAACACUGA---- ..(((.....))).(((---(((((.(((((.......((((((((....--.))))))))........))))-----)...)))).))))...(((.....))).---- ( -27.56) >DroPer_CAF1 50513 96 - 1 AAUCAUGAGUUGGAGCCACGGCAGUCGGUUUAAUUAAUCACCGCUGGGGCACACAGCGGUGCACAA--AAAGCACCAAUGCAACUGCA-ACUGCC------AAUG----- ........(((((.(((.((((.((.((((.....)))))).)))).)))...(((.(((((....--...)))))..(((....)))-.)))))------))).----- ( -31.40) >consensus AAUCAUGAGUUGGAGCU___GCAGUCGGUUUAAUUAAUCACCGCUGUGCC__CCAGCGGUGCGCAA__AAAGC_____UAAAACUGCG_CCAGCCGCAACCACUG_____ .....((((((((.((....))..))))))))......((((((((.......))))))))................................................. (-16.80 = -16.42 + -0.39)


| Location | 11,319,735 – 11,319,829 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -23.27 |
| Energy contribution | -22.80 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11319735 94 - 22407834 ---------AGU-CAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGGAGUU---GCAGUCGGUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCGCAA--AAA ---------...-...(((((((...(((((((.((.....)).)))))))....))---).))))............((((((((....--.)))))))).....--... ( -21.20) >DroPse_CAF1 50131 100 - 1 ---------AGUCCAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGGAGCCACGGCAGUCGGUUUAAUUAAUCACCGCUGGGGCACACAGCGGUGCACAA--AAA ---------.(((...(((((.....(((((((.((.....)).))))))).)))))..)))................((((((((.......)))))))).....--... ( -27.30) >DroEre_CAF1 39073 91 - 1 -------------CAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGCUGGAGCU---GCGCUCGGUUUAAUUAAUCACCGCUGUGUC--CCAGCGGUGCGCAA--AAA -------------...(((((.......(((((.((.....)).)))))...)))))---((((..((((.....))))(((((((....--.)))))))))))..--... ( -24.40) >DroYak_CAF1 37493 103 - 1 CAGUAGUGGAGU-GAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGGAGCU---GCGCUCGAUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCGCAA--AAA .....(((((((-(..(((((.....(((((((.((.....)).))))))).)))))---.)))))............((((((((....--.)))))))))))..--... ( -31.70) >DroAna_CAF1 33303 102 - 1 CG---UUUGCGU-CAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGAAGCU---GCAGUCGGUUUAAUUAAUCACCGCUGUGCC--CCAGCGGUGCACAAAAAAA ((---.((((..-...(((((.....(((((((.((.....)).))))))).)))))---)))).))...........((((((((....--.)))))))).......... ( -25.20) >DroPer_CAF1 50540 100 - 1 ---------AGUCCAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGGAGCCACGGCAGUCGGUUUAAUUAAUCACCGCUGGGGCACACAGCGGUGCACAA--AAA ---------.(((...(((((.....(((((((.((.....)).))))))).)))))..)))................((((((((.......)))))))).....--... ( -27.30) >consensus _________AGU_CAAGGCUUAAAUAAAUUCGUCAUUAUAAAUCAUGAGUUGGAGCU___GCAGUCGGUUUAAUUAAUCACCGCUGUGCC__CCAGCGGUGCACAA__AAA ................(((((.....(((((((.((.....)).))))))).)))))...((....((((.....))))(((((((.......)))))))))......... (-23.27 = -22.80 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:18 2006