| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,312,959 – 11,313,072 |
| Length | 113 |
| Max. P | 0.781055 |

| Location | 11,312,959 – 11,313,072 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.96 |
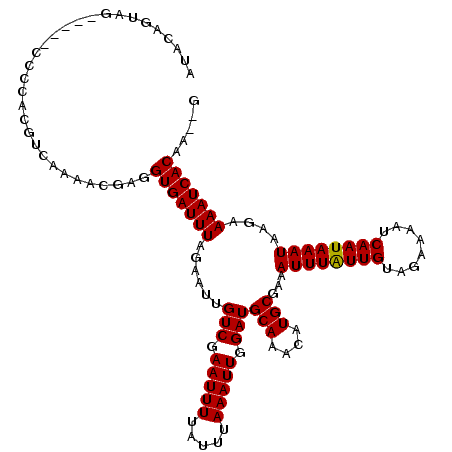
| Structure conservation index | 0.96 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11312959 113 - 22407834 AUACAGUAG-----CCCCACGUCAAAACGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUAUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAA--A .........-----.....(((....)))..(((((((......(((.(((((.....))))).)))(((....)))...((((((((........))))))))....)))))))..--. ( -18.40) >DroVir_CAF1 31473 113 - 1 AUACAGUAG-----CCCCACGUCAAAAUGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUGUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAA--G .........-----.................(((((((......(((.(((((.....))))).)))(((....)))...((((((((........))))))))....)))))))..--. ( -17.60) >DroPse_CAF1 45369 115 - 1 ACACAGUAG-----CCCCACGUCAAAACGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUAUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAAAAA .........-----.....(((....)))..(((((((......(((.(((((.....))))).)))(((....)))...((((((((........))))))))....)))))))..... ( -18.40) >DroGri_CAF1 38421 113 - 1 AUACAGUAG-----CCCCACGUCAAAAUGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUAUUGUUGAAAAUCAAUAAAUAAGAAAAUCACAA--G .........-----.................(((((((......(((.(((((.....))))).)))(((....)))...((((((((.(....).))))))))....)))))))..--. ( -18.00) >DroWil_CAF1 41647 117 - 1 ACACAGUAGUUGCUCCCCACGUCAAAACGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCAAAAUUUAUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAA--- .....(((.(((((((.(((.((.....)).)))...(((((((....)))))))........))).))))...)))...((((((((........)))))))).............--- ( -21.30) >DroMoj_CAF1 45924 111 - 1 --ACAGUAG-----CCCCACGUCAAAAUCAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUAUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAA--G --.......-----.................(((((((......(((.(((((.....))))).)))(((....)))...((((((((........))))))))....)))))))..--. ( -17.60) >consensus AUACAGUAG_____CCCCACGUCAAAACGAGGUGAUUUAGAAUUGUCGAAUUUUAUUUAAAUUGGAUGCAAACAUGCGAAAUUUAUUGUAGAAAAUCAAUAAAUAAGAAAAUCACAA__G ...............................(((((((......(((.(((((.....))))).)))(((....)))...((((((((........))))))))....)))))))..... (-17.74 = -17.60 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:12 2006