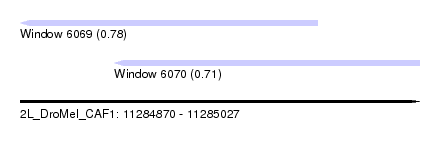
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,284,870 – 11,285,027 |
| Length | 157 |
| Max. P | 0.779734 |

| Location | 11,284,870 – 11,284,987 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
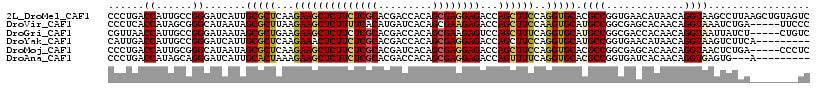
>2L_DroMel_CAF1 11284870 117 - 22407834 CCCUGACCAUUGCCGGGAUCAUUGCGCUCAAGAAGCUCUUCUCGCACGACCACAGCGAGGAGACCAGCUUCCAGGUGCACGCCGGUGAACAUAACAGGUAAGCCUUAAGCUGUAGUC .((((....(..(((((.....((((((...((((((((((((((.........)))))))))...)))))..))))))).))))..)......))))..(((.....)))...... ( -39.80) >DroVir_CAF1 6585 112 - 1 CCCUCACCAUAGCGGGCAUAAUAGCGCUUAAGAAGCUCUUUUCACAUGAUCACAGCGAAGAGACCAGCUUCCAAGUGCAUGCCGGCGAGCACAACAGGUAAAUCUGA-----UUCCC ...........((.(((((....((((((..((((((((((((.(.((....))).)))))))...))))).))))))))))).))........((((....)))).-----..... ( -32.80) >DroGri_CAF1 4816 112 - 1 CGUUAACCAUUGCCGGGAUAAUAGCGCUGAAGAAGCUCUUCUCGCACGACCACAGCGAAGAGUCCAGCUUUCAGGUGCAUGCCGGCGACCACAACAGGUAAUUAUCU-----CUGUC .........(((((((.((....(((((...((((((((..((((.........))))..))...))))))..))))))).))))))).....(((((........)-----)))). ( -34.40) >DroYak_CAF1 4158 108 - 1 CAUUGACCAUUGCCGGGAUCAUUGCGCUCAAGAAACUCUUCUCGCACGACCACAGCGAGGAGACCAGCUUCCAGGUGCAUGCCGGUGAACAUAACAGGUAAGUCUUCA--------- ....(((..(..((((.(....((((((.(((....(((((((((.........)))))))))....)))...))))))).))))..)((.......))..)))....--------- ( -31.50) >DroMoj_CAF1 8751 112 - 1 CCCUGACCAUUGCGGGCAUAAUAGCGCUCAAGAAGCUCUUCUCGCACGAUCACAGCGAGGAGACCAGCUUCCAGGUGCACGCCGGCGAGCACAACAGGUAACUCUGA-----CCCUC .((((....((((.(((......(((((...((((((((((((((.........)))))))))...)))))..)))))..))).))))......)))).........-----..... ( -39.40) >DroAna_CAF1 4097 105 - 1 CCCUGACCAUAGCAGGGAUCAUUGCACUAAAGAAGCUCUUCUCGCACGACCACAGCGAGGAGACCAGUUUUCAGGUGCACGCCGGUGAUCACAACAGGUGAGUG---A--------- (((((.......))))).((((((((((...((((((((((((((.........)))))))))...)))))..))))).((((.((.......)).))))))))---)--------- ( -37.80) >consensus CCCUGACCAUUGCCGGGAUAAUAGCGCUCAAGAAGCUCUUCUCGCACGACCACAGCGAGGAGACCAGCUUCCAGGUGCACGCCGGCGAACACAACAGGUAAGUCUGA________UC ......((......)).......(((((...((((((((((((((.........))))))))...))))))..))))).((((.............))))................. (-25.50 = -25.34 + -0.16)
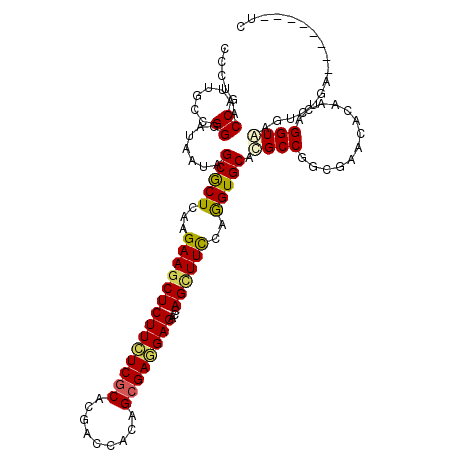

| Location | 11,284,907 – 11,285,027 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -31.69 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
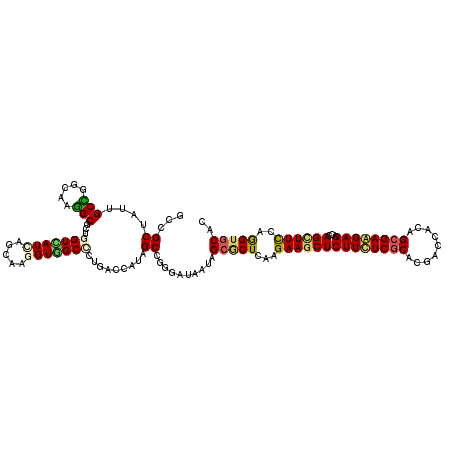
>2L_DroMel_CAF1 11284907 120 - 22407834 GCCGCGAUCGCCGGCAAGGCUCUGGUCAUCAGCAAGGUGGCCCUGACCAUUGCCGGGAUCAUUGCGCUCAAGAAGCUCUUCUCGCACGACCACAGCGAGGAGACCAGCUUCCAGGUGCAC .....((((.(((((((((.((.(((((((.....)))))))..)))).)))))))))))..((((((...((((((((((((((.........)))))))))...)))))..)))))). ( -58.60) >DroVir_CAF1 6617 120 - 1 GCCGCUAUUGCCGGCAAGGCGCUGGUAAUCAGUAAAGUGGCCCUCACCAUAGCGGGCAUAAUAGCGCUUAAGAAGCUCUUUUCACAUGAUCACAGCGAAGAGACCAGCUUCCAAGUGCAU .((((((((((((((.....))))))))).......((((......)))))))))........((((((..((((((((((((.(.((....))).)))))))...))))).)))))).. ( -43.10) >DroGri_CAF1 4848 120 - 1 GCAGCUAUAGCUGGCAAGGCACUGGUCAUCAGCAAAGUGGCGUUAACCAUUGCCGGGAUAAUAGCGCUGAAGAAGCUCUUCUCGCACGACCACAGCGAAGAGUCCAGCUUUCAGGUGCAU (((.((..(((((((...((..((((((((.(((..((((......)))))))...)))....(((..((((.....)))).)))..)))))..)).....).))))))...)).))).. ( -39.80) >DroWil_CAF1 14037 120 - 1 GCAGCAAUUGCUGGUAAAGCAUUGGUAAUCAGCAAGGUUGCUCUAACCAUAGCUGGAAUUAUAGCUCUGAAGAAACUCUUUUCGCAUGAUCACAGCGAAGAGACAAGCUUUCAGGUCCAU (((((..((((((((...((....)).)))))))).))))).........(((((......)))))((((((....(((((((((.((....)))))))))))....).)))))...... ( -35.60) >DroMoj_CAF1 8783 120 - 1 GCCGCUAUUGCCGGCAAGGCGCUUGUGAUUAGCAAGGUCGCCCUGACCAUUGCGGGCAUAAUAGCGCUCAAGAAGCUCUUCUCGCACGAUCACAGCGAGGAGACCAGCUUCCAGGUGCAC (((((....).))))...(((((((......((((((((.....)))).))))((((........))))..((((((((((((((.........)))))))))...)))))))))))).. ( -50.20) >DroAna_CAF1 4122 120 - 1 GCGGCGAUUGCCGGCAAGGCGCUGGUCAUCAGCAAGGUGGCCCUGACCAUAGCAGGGAUCAUUGCACUAAAGAAGCUCUUCUCGCACGACCACAGCGAGGAGACCAGUUUUCAGGUGCAC ...(((((.((((((.....)))))).))).))..((((((((((.......))))).)))))(((((...((((((((((((((.........)))))))))...)))))..))))).. ( -47.10) >consensus GCCGCUAUUGCCGGCAAGGCGCUGGUCAUCAGCAAGGUGGCCCUGACCAUAGCCGGGAUAAUAGCGCUCAAGAAGCUCUUCUCGCACGACCACAGCGAAGAGACCAGCUUCCAGGUGCAC ...((....(((.....)))...(((((((.....))))))).........))..........(((((...((((((((((((((.........))))))))...))))))..))))).. (-31.69 = -31.50 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:01 2006