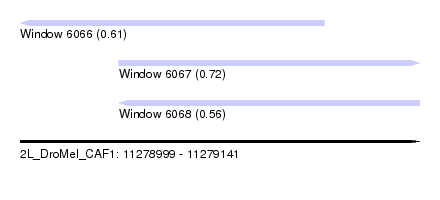
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,278,999 – 11,279,141 |
| Length | 142 |
| Max. P | 0.716798 |

| Location | 11,278,999 – 11,279,107 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -47.42 |
| Consensus MFE | -35.17 |
| Energy contribution | -36.40 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11278999 108 - 22407834 UGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU------CUGCAUCUGCUGGUACUGCUUAACAGUCACCGUCG ..........((((((((((((...((.(((.(((.((((....)))).))).)))))..)))))((((...(------(.((....)).))..))))........))))))). ( -45.10) >DroVir_CAF1 94348 108 - 1 UGUUUUCUGUGAUGGUUGGUGCAUUGGCAGCUG------CAGCUGCCUGUGCAGCAGCCAGGGCGGCGGCCUUUUGCUGUUGCAUUUGCUGAAACUGUUUGAUAGUAACAGUUG .....((.(..(((((((.(((((.((((((..------..)))))).))))).))))))..((((((((.....))))))))..)..).))(((((((.......))))))). ( -50.00) >DroSec_CAF1 111149 108 - 1 UGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU------CUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCG ..........((((((((.(((...((..((((.(.(((.(((((...(((((((.((....)).))))))).------))))))))).))))...))...))).)))))))). ( -49.70) >DroSim_CAF1 119190 108 - 1 UGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAGUGCCGCAGCCUU------CUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCG ..........((((((((.(((......(((.(((.((((....)))).))).)))((.((((((((((....------......)))).))))))))...))).)))))))). ( -49.90) >DroEre_CAF1 113417 108 - 1 UGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGACGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU------CUGCAUCUGCUGGUACUGCUUAACGGUUACCGUCG ..........((((((((.(((...((..((((....(((.((((...(((((((.((....)).))))))).------)))).)))..))))...))...))).)))))))). ( -47.20) >DroYak_CAF1 115278 108 - 1 UGUUCUCCGUAAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU------CUGCAUCUGCUGGUACUGCUUAACGGUCACAGUCG .((((.((.....)).))))..(((((.((((((..((((.((((...(((((((.((....)).))))))).------))))(((....)))...)))).)))))))))).). ( -42.60) >consensus UGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU______CUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCG ..........((((((((.(((...((..((((....((.(((((...(((((((.((....)).))))))).......))))).))..))))...))...))).)))))))). (-35.17 = -36.40 + 1.23)

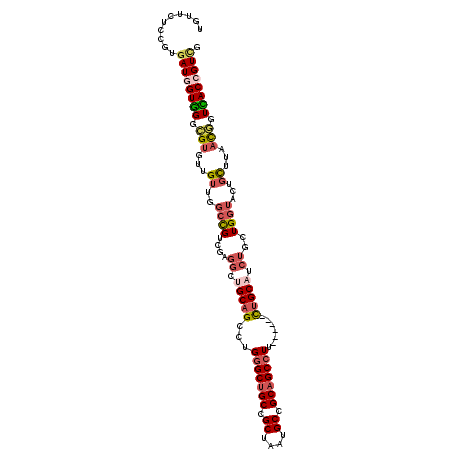
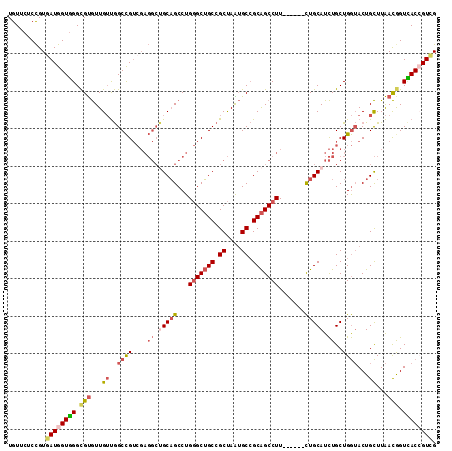
| Location | 11,279,034 – 11,279,141 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -19.77 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11279034 107 + 22407834 -AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCAACAACACGCCCACCAUCACGGAGAACAUCAUUAUCCAGCAGCCCACUUCGACUACGGCGAC -..((((((.((....)).))))))..((((((.(((.....))).....................(((...........))).))))))....(((.(....)))). ( -37.60) >DroVir_CAF1 94388 102 + 1 AAAGGCCGCCGCCCUGGCUGCUGCACAGGCAGCUG------CAGCUGCCAAUGCACCAACCAUCACAGAAAACAUCAUCAUACAGCAACCCACAUCAACGACAGCCGU ...(((....)))..((((((((((..((((((..------..))))))..))))............((.....)).......................).))))).. ( -27.40) >DroSim_CAF1 119225 107 + 1 -AAGGCUGCGGCACUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCAACAACACGCCCACCAUCACGGAGAACAUCAUUAUCCAGCAGCCCACUUCGACUACGGCGAC -..((((((.((....)).))))))..((((((.(((.....))).....................(((...........))).))))))....(((.(....)))). ( -37.60) >DroEre_CAF1 113452 107 + 1 -AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCGUCCUCGACGGCCAACAACACGCCCACCAUCACGGAGAACAUCAUUAUCCAGCAGCCCACUUCGACUACAGCGAC -..((((((.((....)).))))))..(((((((((...)))(((.........))).........(((...........))).))))))....(((.(....)))). ( -34.30) >DroYak_CAF1 115313 107 + 1 -AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCAACAACACGCCCACCAUUACGGAGAACAUCAUUAUCCAGCAACCCACUUCGACAACGGCGGC -..((((((.((....)).))))))..((((((......).))))).......((((........(((((......................))))).....)))).. ( -31.47) >DroAna_CAF1 106801 101 + 1 -AAGGCGGCGGCUUUGGCGGCAGCACAAGCGGCAGCAGCA------GCAAACACACCCACAAUCACGGAAAACAUCAUUAUCCAGCAGCCCACUUCAACUUCCACAAC -..(((.((.(((...((.((.((....)).)).))...)------))..................(((...........))).)).))).................. ( -20.30) >consensus _AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCAACAACACGCCCACCAUCACGGAGAACAUCAUUAUCCAGCAGCCCACUUCGACUACGGCGAC ...(((((((((....((.(((((....))))).)).......)))....................(((...........))).)))))).................. (-19.77 = -21.72 + 1.94)

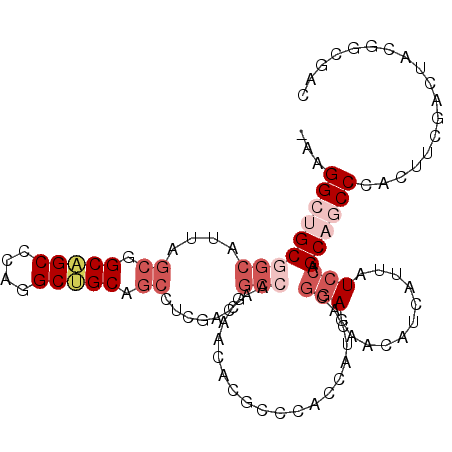
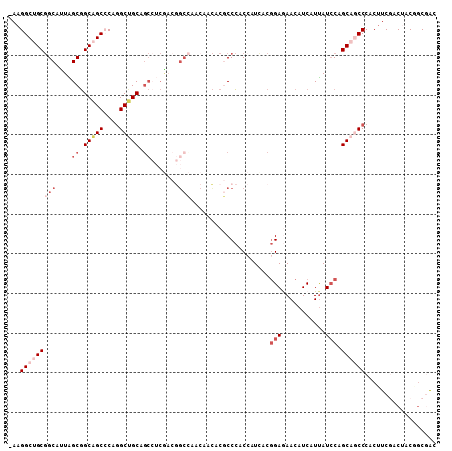
| Location | 11,279,034 – 11,279,141 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -27.91 |
| Energy contribution | -29.92 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11279034 107 - 22407834 GUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU- .((((....).)))...(((((((..((((((.((((((.((.....)).))))))))))))((((.....)))))))))))(((((((.((....)).))))))).- ( -45.30) >DroVir_CAF1 94388 102 - 1 ACGGCUGUCGUUGAUGUGGGUUGCUGUAUGAUGAUGUUUUCUGUGAUGGUUGGUGCAUUGGCAGCUG------CAGCUGCCUGUGCAGCAGCCAGGGCGGCGGCCUUU ..((((((((((...(..((..((...........))..))..)..((((((.(((((.((((((..------..)))))).))))).)))))).))))))))))... ( -49.70) >DroSim_CAF1 119225 107 - 1 GUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAGUGCCGCAGCCUU- .((((....).)))...(((((((..((((((.((((((.((.....)).))))))))))))((((.....)))))))))))(((((((.((....)).))))))).- ( -45.30) >DroEre_CAF1 113452 107 - 1 GUCGCUGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGACGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU- ..((((........))))((((((.(((((....)))))(((.(((((((.(((.....))).))))))).))).)))))).(((((((.((....)).))))))).- ( -48.30) >DroYak_CAF1 115313 107 - 1 GCCGCCGUUGUCGAAGUGGGUUGCUGGAUAAUGAUGUUCUCCGUAAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU- .................(((((((..((((((.((((((.((.....)).))))))))))))((((.....)))))))))))(((((((.((....)).))))))).- ( -42.60) >DroAna_CAF1 106801 101 - 1 GUUGUGGAAGUUGAAGUGGGCUGCUGGAUAAUGAUGUUUUCCGUGAUUGUGGGUGUGUUUGC------UGCUGCUGCCGCUUGUGCUGCCGCCAAAGCCGCCGCCUU- ...((((...........((((..((((((((.(((.....))).))))).((((..(..((------.((....)).))..)..).))).))).)))).))))...- ( -26.90) >consensus GUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCGCAGCCUU_ .................(((((((.(((.(((...))).))).(((((((.(((.....))).))))))).....)))))))(((((((.((....)).))))))).. (-27.91 = -29.92 + 2.00)

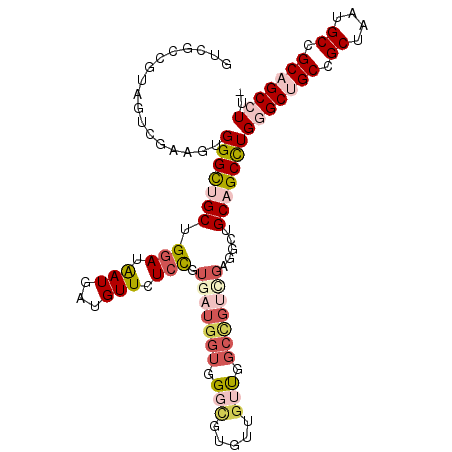
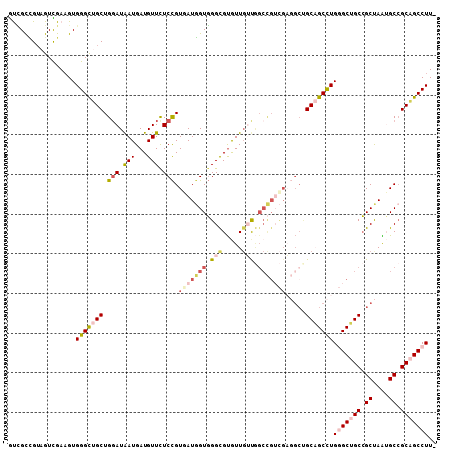
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:00 2006