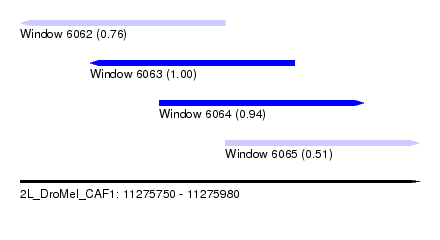
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,275,750 – 11,275,980 |
| Length | 230 |
| Max. P | 0.995639 |

| Location | 11,275,750 – 11,275,868 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.82 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
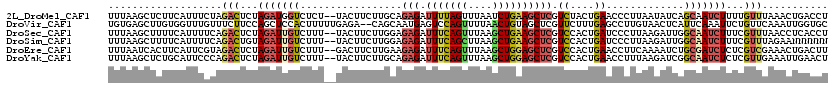
>2L_DroMel_CAF1 11275750 118 - 22407834 UUUAAGCUCUUCAUUUCUAGACUCUAGAUGGUCUCU--UACUUCUUGCAGAGAUUUUAGUUUAAUCUGAAGCUCGUCUACUGAACCCUUAAUAUCAGCAAUCUUUUGUUUAAACUGACCU (((((((...........(((((......)))))..--......((((.(((.((((((......))))))))).((....)).............))))......)))))))....... ( -17.20) >DroVir_CAF1 91362 118 - 1 UGUGAGCUUGUGGUUUGUUUCUUCCAGCUCCACUUUUUGAGA--CAGCAAUGAGUCCAGUUUUAACUGUAGCUCGUUCUUUGAGCCUUGUAACUCAUUCAAAUUCUGUUCAAAUUGGUGC ...(((((.(.((.......)).).)))))((((.((((((.--(((.(((((((.((((....))))..(((((.....)))))......)))))))......)))))))))..)))). ( -29.00) >DroSec_CAF1 107841 118 - 1 UUUAAGCUUUUCAUUUUCAGACUCUAGAUUGUCUUU--UACUUCUUGGAGAGAUUUCAGUUUAAGCUGAAGCUCGUCCACUGAUCCCUUAAGAUUGGCAAUCUUUCGUUUAACCUCACCU ..................((((...((((((((...--.......(((((((.(((((((....)))))))))).))))..((((......))))))))))))...)))).......... ( -27.70) >DroSim_CAF1 115879 118 - 1 UUUAAGCUUUUCAUUUUCAGACUGUAGAUUGUCUUU--UACUUCUUGGAGAGAUUUCAGCUUAAGCUGAAGCUCGUCCACUGAUCCCUUAAGAUUGGCAAUCUUUCGUUUAGAANNNNNN ...............(((((((...((((((((...--.......(((((((.(((((((....)))))))))).))))..((((......))))))))))))...)))).)))...... ( -30.50) >DroEre_CAF1 110260 118 - 1 UUUAAUCACUUCAUUCGUAGACUCUAGAUUGUCUUU--GACUUCUUGAAGAGAUUUCAGUUUAAGCUGGAGCUCGUCCACUGAACCUUCAAAAUCUGCGAUCUCUCGUCGAAACUGACUU ...((((.(((((.(((.((((........)))).)--)).....))))).))))(((((((..((.((((.((((....(((....)))......)))).)))).))..)))))))... ( -29.80) >DroYak_CAF1 111925 118 - 1 UUUAAGCUCUGCAUUCCCAGACUCUAGAUUGUCUUU--UACUUCUUGCAGAGAUUUCAGUUUAAGCUGGAGCUCGUCCACUGAACCUUUAAGAUCGGCAAUCUCUCGUUGAAAUUGAACU ......(((((((.....((((........))))..--.......)))))))..((((((((.(((.((((...(((..((.........))...)))...)))).))).)))))))).. ( -26.14) >consensus UUUAAGCUCUUCAUUUCCAGACUCUAGAUUGUCUUU__UACUUCUUGCAGAGAUUUCAGUUUAAGCUGAAGCUCGUCCACUGAACCCUUAAGAUCGGCAAUCUUUCGUUUAAACUGACCU ...................(((...(((((((.................(((.(((((((....)))))))))).((....)).............)))))))...)))........... (-10.09 = -10.82 + 0.73)

| Location | 11,275,790 – 11,275,908 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11275790 118 - 22407834 CCACCAUGUUCUGUAGUUCCUGAGCAUCUUUUAGCUUUCUUUUAAGCUCUUCAUUUCUAGACUCUAGAUGGUCUCU--UACUUCUUGCAGAGAUUUUAGUUUAAUCUGAAGCUCGUCUAC ......(((...(.(((....((.(((((....((((......))))(((........)))....))))).))...--.))).)..)))(((.((((((......)))))))))...... ( -21.60) >DroGri_CAF1 132043 118 - 1 CCAAAUCACUCUGCAUAUUUAGGACAUUUUUCAGUUUUACUGUCAGCUGCUGAUUCGCUGUCUCCAGCUCCAGUUUAUUAGU--GAGCAAUGAGUCCAGCUGCAACUGCAGUUCAUUCUU ..........(((((....((((((........)))))).((.((((((..(((((((((....))))....(((((....)--))))...)))))))))))))..)))))......... ( -29.80) >DroSec_CAF1 107881 118 - 1 CCACUUUGUUCUGUAGAUUCUGAGCAUCAUUUAGCUUUAUUUUAAGCUUUUCAUUUUCAGACUCUAGAUUGUCUUU--UACUUCUUGGAGAGAUUUCAGUUUAAGCUGAAGCUCGUCCAC ...........(.((((.((((((........(((((......))))).......)))))).)))).)........--.......(((((((.(((((((....)))))))))).)))). ( -29.16) >DroSim_CAF1 115919 118 - 1 CCACUUUGUCCUGUAGAUUCUGAGCGUCUUUUAGCUUUAUUUUAAGCUUUUCAUUUUCAGACUGUAGAUUGUCUUU--UACUUCUUGGAGAGAUUUCAGCUUAAGCUGAAGCUCGUCCAC ..........((((((..((((((.((.....(((((......)))))....)).)))))))))))).........--.......(((((((.(((((((....)))))))))).)))). ( -29.70) >DroEre_CAF1 110300 109 - 1 CCACCAGGU---------CUUGAGCAUCCUUUAGCUUACUUUUAAUCACUUCAUUCGUAGACUCUAGAUUGUCUUU--GACUUCUUGAAGAGAUUUCAGUUUAAGCUGGAGCUCGUCCAC ......((.---------..(((((.(((...((((((.....((((.(((((.(((.((((........)))).)--)).....))))).))))......)))))))))))))).)).. ( -30.80) >DroYak_CAF1 111965 118 - 1 CCACCAUGUUUUGUAGAUCCUGAGCUUGUUUUAGCUUACCUUUAAGCUCUGCAUUCCCAGACUCUAGAUUGUCUUU--UACUUCUUGCAGAGAUUUCAGUUUAAGCUGGAGCUCGUCCAC ....................((..(..(((((((((((.((..((.(((((((.....((((........))))..--.......))))))).))..))..)))))))))))..)..)). ( -28.14) >consensus CCACCAUGUUCUGUAGAUCCUGAGCAUCUUUUAGCUUUAUUUUAAGCUCUUCAUUCCCAGACUCUAGAUUGUCUUU__UACUUCUUGCAGAGAUUUCAGUUUAAGCUGAAGCUCGUCCAC .....................((((....((((((((.........(((((((.....((((........))))...........)))))))..........))))))))))))...... (-13.96 = -14.68 + 0.73)

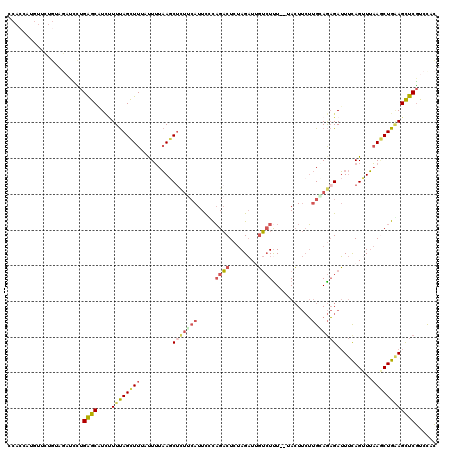
| Location | 11,275,830 – 11,275,948 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -11.69 |
| Energy contribution | -13.42 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11275830 118 + 22407834 UA-AGAGACCAUCUAGAGUCUAGAAAUGAAGAGCUUAAAAGAAAGCUAAAAGAUGCUCAGGAACUACAGAACAUGGUGGAUAAGGAGCGAAAGCUCAAUUCCUCUCUACGAGAAGAUUU ..-(((((((((((..(((((.((.((....(((((......))))).....))..)).)).)))..)))...))))(((....((((....))))...))).))))............ ( -29.00) >DroSec_CAF1 107921 118 + 1 UA-AAAGACAAUCUAGAGUCUGAAAAUGAAAAGCUUAAAAUAAAGCUAAAUGAUGCUCAGAAUCUACAGAACAAAGUGGAUAAGGAGCAAAAGCUCAAUUCUUCUCUACGAGAAGAUUU ..-............((((............(((((......)))))......(((((...((((((........))))))...)))))...))))...(((((((...)))))))... ( -28.90) >DroSim_CAF1 115959 118 + 1 UA-AAAGACAAUCUACAGUCUGAAAAUGAAAAGCUUAAAAUAAAGCUAAAAGACGCUCAGAAUCUACAGGACAAAGUGGAUAAGGAGCAAAAGCUCAAUUCCUCUCUACGAGAAGAUUU ..-..((((........)))).....(((..(((((......))))).......((((...((((((........))))))...))))......)))..((.((((...)))).))... ( -23.90) >DroEre_CAF1 110340 109 + 1 UC-AAAGACAAUCUAGAGUCUACGAAUGAAGUGAUUAAAAGUAAGCUAAAGGAUGCUCAAG---------ACCUGGUGGAUAAAGAGCGAAAGCUUAAUGCCUCUCUGCAAAAUGAUUU ((-(..((((.(((..((..(((.....((....))....)))..))..))).)).)).((---------(...((((......((((....))))..))))..)))......)))... ( -18.90) >DroYak_CAF1 112005 118 + 1 UA-AAAGACAAUCUAGAGUCUGGGAAUGCAGAGCUUAAAGGUAAGCUAAAACAAGCUCAGGAUCUACAAAACAUGGUGGAUAAGGAGCGAAAGCUUAAUGCCUCUCUACGAGAUGAUUU ..-.....((.(((((((((((((..((...((((((....))))))....))..))))))((((((........))))))...((((....)))).......))))).))..)).... ( -31.10) >DroAna_CAF1 103445 119 + 1 CAGAAACAGGAACUGGAGGCCGUAAACGAGGAGAUGAAGUUGAAUUUAAAGGCAUCUCAGCAUCUACAGGCCCAGCUGGAGGAGGAGCGAAAAGUAGUUGCUGAUCUACAAGAAUCCUA .......((((.(..(.(((((((...(..((((((...(((....)))...))))))..)...))).))))...)..).(((..(((((.......)))))..))).......)))). ( -31.50) >consensus UA_AAAGACAAUCUAGAGUCUGAAAAUGAAGAGCUUAAAAGAAAGCUAAAAGAUGCUCAGGAUCUACAGAACAAGGUGGAUAAGGAGCGAAAGCUCAAUGCCUCUCUACGAGAAGAUUU .............(((((...(((.......(((((......))))).......((((...((((((........))))))...))))..........)))..)))))........... (-11.69 = -13.42 + 1.73)

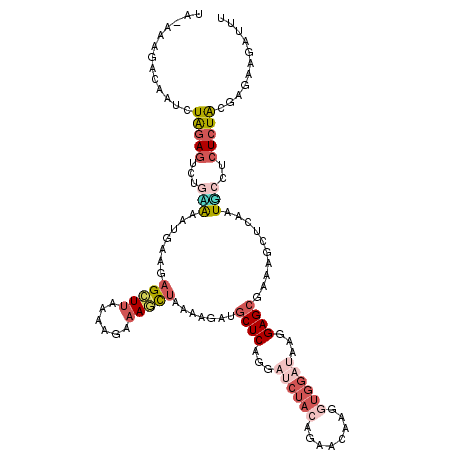
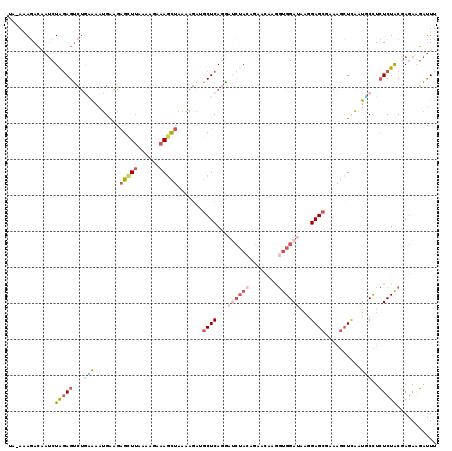
| Location | 11,275,868 – 11,275,980 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -10.60 |
| Energy contribution | -13.85 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.40 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11275868 112 + 22407834 AGAAAGCUAAAAGAUGCUCAGGAACUACAGAACAUGGUGGAUAAGGAGCGAAAGCUCAAUUCCUCUCUACGAGAAGAUUUUGACAAACUGGAACAAACCAAACU---CGAU----CUCG (((.(((........))).(((((((((........)))).....((((....))))..))))).))).(((((.((.((((................)))).)---)..)----)))) ( -25.59) >DroSec_CAF1 107959 112 + 1 AUAAAGCUAAAUGAUGCUCAGAAUCUACAGAACAAAGUGGAUAAGGAGCAAAAGCUCAAUUCUUCUCUACGAGAAGAUUUGGGCAAACUGGAACAAACCAAACU---CGGU----CUCG ......(((.....(((((...((((((........))))))...)))))...((((((.(((((((...))))))).))))))....))).....(((.....---.)))----.... ( -32.70) >DroSim_CAF1 115997 112 + 1 AUAAAGCUAAAAGACGCUCAGAAUCUACAGGACAAAGUGGAUAAGGAGCAAAAGCUCAAUUCCUCUCUACGAGAAGAUUUGGGCAAACUGGAAAAAACCAAACU---CGAU----CUCG ...............((((...((((((........))))))...))))....((((((.((.((((...)))).)).))))))....(((......)))....---....----.... ( -27.70) >DroEre_CAF1 110378 103 + 1 AGUAAGCUAAAGGAUGCUCAAG---------ACCUGGUGGAUAAAGAGCGAAAGCUUAAUGCCUCUCUGCAAAAUGAUUUGGGCAAACUAGGACAAAUAAAAAC---GGAU----CUCG .((.(((........)))....---------.((((((.......((((....))))..(((((.((........))...))))).))))))))..........---....----.... ( -23.20) >DroYak_CAF1 112043 112 + 1 GGUAAGCUAAAACAAGCUCAGGAUCUACAAAACAUGGUGGAUAAGGAGCGAAAGCUUAAUGCCUCUCUACGAGAUGAUUUGGGCAAACUGGAACAAACACAAAC---UGAU----CUCG ((((((((.......((((...((((((........))))))...))))...))))...))))......((((((.(((((..................)))).---).))----)))) ( -27.07) >DroAna_CAF1 103484 116 + 1 UUGAAUUUAAAGGCAUCUCAGCAUCUACAGGCCCAGCUGGAGGAGGAGCGAAAAGUAGUUGCUGAUCUACAAGAAUCCUACUCCAAUCUGAAA---ACUAAAUUAAAGGAUAUGACUCG ...((((((..(((................)))(((.(((((.((((.......((((........)))).....)))).)))))..)))...---..))))))............... ( -20.99) >consensus AGAAAGCUAAAAGAUGCUCAGGAUCUACAGAACAAGGUGGAUAAGGAGCGAAAGCUCAAUGCCUCUCUACGAGAAGAUUUGGGCAAACUGGAACAAACCAAACU___CGAU____CUCG ......(((......((((...((((((........))))))...))))....((((((.((.((((...)))).)).))))))....)))............................ (-10.60 = -13.85 + 3.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:57 2006