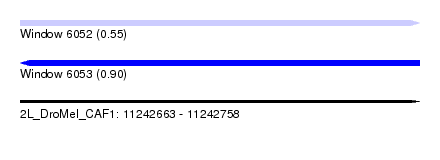
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,242,663 – 11,242,758 |
| Length | 95 |
| Max. P | 0.904413 |

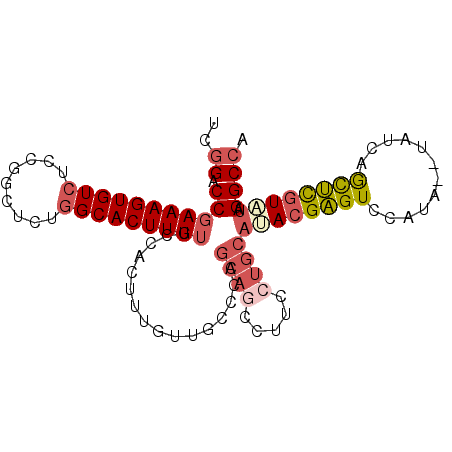
| Location | 11,242,663 – 11,242,758 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -16.62 |
| Energy contribution | -19.10 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11242663 95 + 22407834 UGGCCUAUACGAGCUGAUA--UAUGGACUCGUAUGCAUGAAGGCUGCUGGGCAACAAAGUGGCAAAAGUGCUAGAGACGGAGACACUUUCGGUCCGA .((((.(((((((((....--...)).)))))))((((....((..((.(....)..))..))....))))..((((.(....)..))))))))... ( -30.00) >DroSec_CAF1 74698 97 + 1 UGGCCUAUACGAGCUGAUAUAUAUGGACUCGUAUGCAGGAAGGCUGCUGGGCAACAAAGUGGCAAAAGUGCCAGAGCCGGAGACACUUUCGGUCCGA ..(((((((((((((.........)).)))))))((((.....)))).)))).......(((((....)))))(..((((((....))))))..).. ( -32.80) >DroSim_CAF1 78487 86 + 1 UGGCCUAUACGAGCUGAUAUAUAUGGACUCGUAUGCAGGAAGGCUGCUGGGC-----------AAAAGUGCCAGAGCCGGAGACACUUUCGGUCCGA .((((((((((((((.........)).))))))))..((((((((.((((.(-----------(....))))))))))(....)..))))))))... ( -30.00) >DroEre_CAF1 75710 95 + 1 UGGCCUAUACGAGUAGAUA--UAUGUACUCGUCUGCAGGAAGGCUGCUGGGCAACAAAGUGGCAAAAGUGCCAGAACCGGAGACACUUUCGGUCCGA ..(((((.(((((((.(..--..).)))))))..((((.....))))))))).......(((((....)))))(.(((((((....))))))).).. ( -32.30) >DroWil_CAF1 103024 84 + 1 UUGCCUAUAUACACUACUA--C-UGAACACUUGGGCAUAAA---------GUAGCAAAGUGGCAAAAGUGCCGACACUGGAGACACUUU-GGUCCCA .((((((............--.-........))))))....---------....(((((((.....((((....)))).....))))))-)...... ( -18.70) >DroYak_CAF1 78593 95 + 1 UGGCCUAAACGAGUAGAUA--UAUGUACUCGUCUGCAGGAAGGCUGCUGGGCAACAAAGUGGCAAAAGUGCCAGAACCGGAGACACUUUCGGUCCGA ..(((((.(((((((.(..--..).)))))))..((((.....))))))))).......(((((....)))))(.(((((((....))))))).).. ( -32.60) >consensus UGGCCUAUACGAGCUGAUA__UAUGGACUCGUAUGCAGGAAGGCUGCUGGGCAACAAAGUGGCAAAAGUGCCAGAACCGGAGACACUUUCGGUCCGA ..(((((((((((.(.((....)).).)))))).((((.....))))))))).......(((((....)))))(.(((((((....))))))).).. (-16.62 = -19.10 + 2.48)

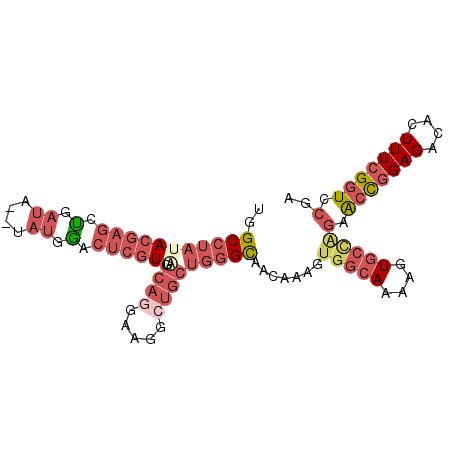
| Location | 11,242,663 – 11,242,758 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11242663 95 - 22407834 UCGGACCGAAAGUGUCUCCGUCUCUAGCACUUUUGCCACUUUGUUGCCCAGCAGCCUUCAUGCAUACGAGUCCAUA--UAUCAGCUCGUAUAGGCCA ..((.((((((((((...........))))))))((......(((((...)))))......))((((((((.....--.....)))))))).)))). ( -24.70) >DroSec_CAF1 74698 97 - 1 UCGGACCGAAAGUGUCUCCGGCUCUGGCACUUUUGCCACUUUGUUGCCCAGCAGCCUUCCUGCAUACGAGUCCAUAUAUAUCAGCUCGUAUAGGCCA ..(((((....).))))..((((.(((((....)))))............((((.....))))((((((((..((....))..)))))))).)))). ( -29.80) >DroSim_CAF1 78487 86 - 1 UCGGACCGAAAGUGUCUCCGGCUCUGGCACUUUU-----------GCCCAGCAGCCUUCCUGCAUACGAGUCCAUAUAUAUCAGCUCGUAUAGGCCA ..(((((....).))))..((((..((((....)-----------)))..((((.....))))((((((((..((....))..)))))))).)))). ( -28.40) >DroEre_CAF1 75710 95 - 1 UCGGACCGAAAGUGUCUCCGGUUCUGGCACUUUUGCCACUUUGUUGCCCAGCAGCCUUCCUGCAGACGAGUACAUA--UAUCUACUCGUAUAGGCCA ..((..((((((((((.........))))))))))))........(((..((((.....))))..(((((((....--....)))))))...))).. ( -29.30) >DroWil_CAF1 103024 84 - 1 UGGGACC-AAAGUGUCUCCAGUGUCGGCACUUUUGCCACUUUGCUAC---------UUUAUGCCCAAGUGUUCA-G--UAGUAGUGUAUAUAGGCAA ((((((.-.....))).))).((((((((....))))((.(((((((---------(..((((....))))..)-)--)))))).)).....)))). ( -25.00) >DroYak_CAF1 78593 95 - 1 UCGGACCGAAAGUGUCUCCGGUUCUGGCACUUUUGCCACUUUGUUGCCCAGCAGCCUUCCUGCAGACGAGUACAUA--UAUCUACUCGUUUAGGCCA ..((..((((((((((.........))))))))))))........(((..((((.....))))(((((((((....--....))))))))).))).. ( -31.10) >consensus UCGGACCGAAAGUGUCUCCGGCUCUGGCACUUUUGCCACUUUGUUGCCCAGCAGCCUUCCUGCAUACGAGUCCAUA__UAUCAGCUCGUAUAGGCCA ..((.(((((((((((.........)))))))))................((((.....))))((((((((............)))))))).)))). (-18.16 = -19.22 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:45 2006