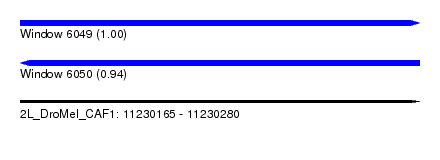
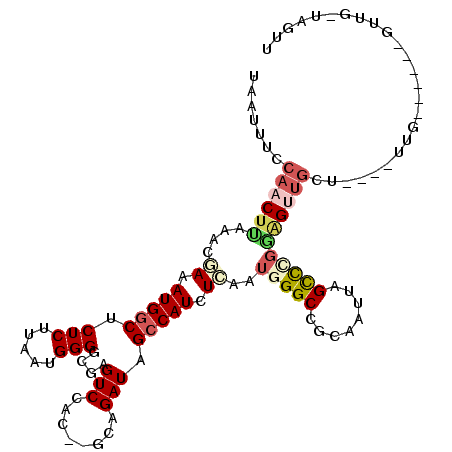
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,230,165 – 11,230,280 |
| Length | 115 |
| Max. P | 0.997328 |

| Location | 11,230,165 – 11,230,280 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -34.21 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11230165 115 + 22407834 AACUAUCAACAAGCGGCAACAAUAGCAACCUCAGGCUAAUUGCGGCCCAUUGAGAUGGCUAUCGGCUAGUGGACUCGCCCCAUUAAGAGAGCCAUUUUGCUGAAGUUGAAAAUUA .....(((((.((((((..(((((((........))).))))..)))....(((((((((.((...((((((.......)))))).)).))))))))))))...)))))...... ( -38.20) >DroPse_CAF1 119922 88 + 1 -----------------------UCCAGCUCCGGGCUAAUUGCGGCCCAUUGAGAUGGCUAUCAG----CGGACUCGCCCCAUUAAGAGAGCCAUUUCGUCUAAGUUGGAAAUUA -----------------------(((((((..(((((......)))))...(((((((((.((.(----((....)))........)).))))))))).....)))))))..... ( -32.30) >DroWil_CAF1 88055 101 + 1 AUGGACCAACU-----CAA----AGAAAC---CAACUAAUUGCGGCCCAUUCAGAUGCCUAUCUGU--ACCGACCCGCCCCAUUAAGAGCGUCAUUUGGUUUAAGUUGGAAAUUA .....((((((-----...----..((((---(((......((((......(((((....))))).--......))))........(.....)..))))))).))))))...... ( -21.82) >DroYak_CAF1 65707 115 + 1 AACUAUCAACUAGCGGCAACAAUAGCCACCUCGGGCUAAUUGCGGCCCAUUGAGAUGGCUAUCGGCUAGUGGACUCGCCCCAUUAAGAGAGCCAUUUUGGUGAAGUUGGAAAUUA .....((((((...(((..((((((((......)))).))))..)))(((..((((((((.((...((((((.......)))))).)).))))))))..))).))))))...... ( -44.60) >DroAna_CAF1 57734 110 + 1 AACUACCAACC-----CAGCCACAACAACUCCCAGCUAAUUGCGGCCCAUUGAGAUGGCUAUCUGCGAGUUGACUCGACCCAUUAAGAGGGCCAUUUCAGCGGAGUUGGAAAUUA .....((((((-----(.(((.(((..............))).)))...(((((((((((.(((.((((....))))........))).))))))))))).)).)))))...... ( -36.04) >DroPer_CAF1 83537 88 + 1 -----------------------UCCAGCUCCGGGCUAAUUGCGGCCCAUUGAGAUGGCUAUCAG----CGGACUCGCCCCAUUAAGAGAGCCAUUUCGUCUAAGUUGGAAAUUA -----------------------(((((((..(((((......)))))...(((((((((.((.(----((....)))........)).))))))))).....)))))))..... ( -32.30) >consensus AACUA_CAAC______CAA____AGCAACUCCGGGCUAAUUGCGGCCCAUUGAGAUGGCUAUCAGC__GCGGACUCGCCCCAUUAAGAGAGCCAUUUCGUCGAAGUUGGAAAUUA .........................((((....((((......))))...((((((((((.((.......((......))......)).)))))))))).....))))....... (-15.33 = -15.92 + 0.59)

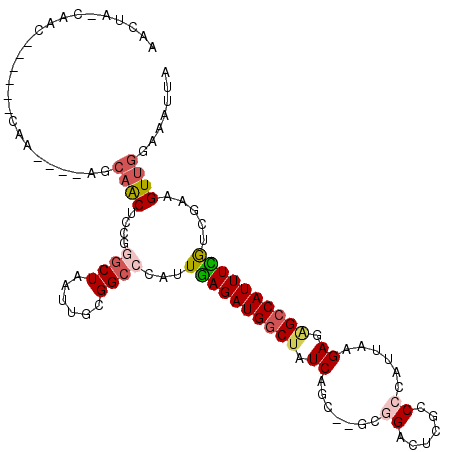
| Location | 11,230,165 – 11,230,280 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11230165 115 - 22407834 UAAUUUUCAACUUCAGCAAAAUGGCUCUCUUAAUGGGGCGAGUCCACUAGCCGAUAGCCAUCUCAAUGGGCCGCAAUUAGCCUGAGGUUGCUAUUGUUGCCGCUUGUUGAUAGUU ........(((((((((((..((((...........(((.((....)).)))((((((.((((((...(((........))))))))).))))))...)))).))))))).)))) ( -35.80) >DroPse_CAF1 119922 88 - 1 UAAUUUCCAACUUAGACGAAAUGGCUCUCUUAAUGGGGCGAGUCCG----CUGAUAGCCAUCUCAAUGGGCCGCAAUUAGCCCGGAGCUGGA----------------------- .....((((.(((....((.(((((((((.....)))(((....))----)....)))))).))..(((((........)))))))).))))----------------------- ( -27.40) >DroWil_CAF1 88055 101 - 1 UAAUUUCCAACUUAAACCAAAUGACGCUCUUAAUGGGGCGGGUCGGU--ACAGAUAGGCAUCUGAAUGGGCCGCAAUUAGUUG---GUUUCU----UUG-----AGUUGGUCCAU ......((((((((((((((.(((((((((....))))))((((.((--.(((((....))))).)).))))....))).)))---))....----)))-----))))))..... ( -35.90) >DroYak_CAF1 65707 115 - 1 UAAUUUCCAACUUCACCAAAAUGGCUCUCUUAAUGGGGCGAGUCCACUAGCCGAUAGCCAUCUCAAUGGGCCGCAAUUAGCCCGAGGUGGCUAUUGUUGCCGCUAGUUGAUAGUU ........(((((((.......(((((.(((....))).))))).(((((((((((((((((((...((((........))))))))))))))))).....))))))))).)))) ( -41.70) >DroAna_CAF1 57734 110 - 1 UAAUUUCCAACUCCGCUGAAAUGGCCCUCUUAAUGGGUCGAGUCAACUCGCAGAUAGCCAUCUCAAUGGGCCGCAAUUAGCUGGGAGUUGUUGUGGCUG-----GGUUGGUAGUU .((((.(((((((((.(((.(((((((.......))(.((((....))))).....))))).))).))(((((((((.(((.....)))))))))))))-----)))))).)))) ( -39.80) >DroPer_CAF1 83537 88 - 1 UAAUUUCCAACUUAGACGAAAUGGCUCUCUUAAUGGGGCGAGUCCG----CUGAUAGCCAUCUCAAUGGGCCGCAAUUAGCCCGGAGCUGGA----------------------- .....((((.(((....((.(((((((((.....)))(((....))----)....)))))).))..(((((........)))))))).))))----------------------- ( -27.40) >consensus UAAUUUCCAACUUAAACGAAAUGGCUCUCUUAAUGGGGCGAGUCCAC__GCAGAUAGCCAUCUCAAUGGGCCGCAAUUAGCCCGGAGUUGCU____UUG______GUUG_UAGUU .......((((((....((.(((((.(((.....)))....(((........))).))))).))..(((((........)))))))))))......................... (-13.65 = -14.60 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:42 2006