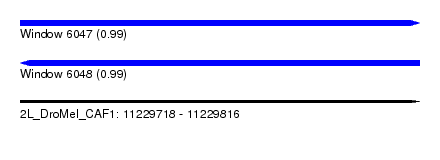
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,229,718 – 11,229,816 |
| Length | 98 |
| Max. P | 0.992966 |

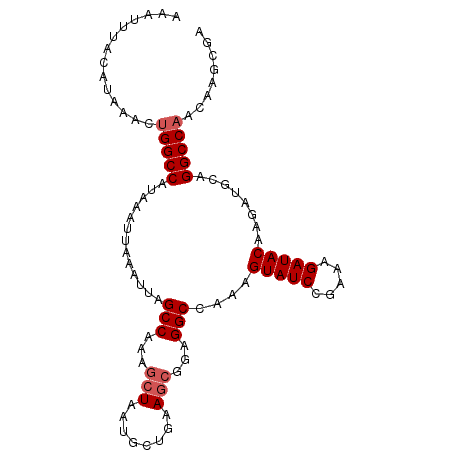
| Location | 11,229,718 – 11,229,816 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
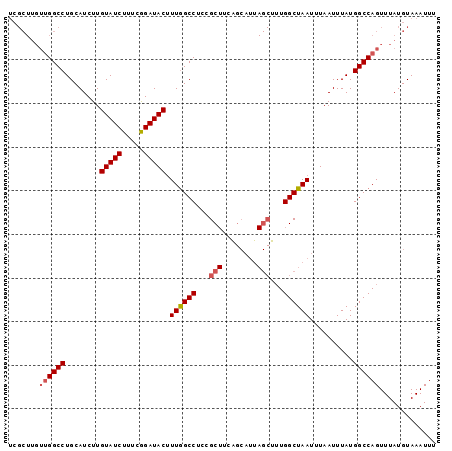
>2L_DroMel_CAF1 11229718 98 + 22407834 UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACGUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU .......((((((........(((((.....)))))(((((((...(((........)))...))))))).........))))))............. ( -25.90) >DroPse_CAF1 119528 75 + 1 ---------GGCCUGCAUCUUGUAUCUGUUAGAUACUUUAGCCGUCGC--------------UGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU ---------((((........(((((.....))))).(((((((....--------------)))))))..........))))............... ( -20.00) >DroSec_CAF1 61881 98 + 1 UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU ..(((....))).(((((...(((((.....))))).((((((...((....))((((((....)))))).........))))))...)))))..... ( -24.50) >DroSim_CAF1 65692 98 + 1 UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU ..(((....))).(((((...(((((.....))))).((((((...((....))((((((....)))))).........))))))...)))))..... ( -24.50) >DroEre_CAF1 62671 98 + 1 UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUUCGCUGCGGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU .(((.((..((((........(((((.....)))))...))))..))..)))((((.((((..(((((..........))))))))).))))...... ( -25.40) >DroYak_CAF1 65270 98 + 1 UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCGGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU ..(((....))).(((((...(((((.....))))).((((((...((....))((((((....)))))).........))))))...)))))..... ( -27.60) >consensus UCGCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUU .......((((((........(((((.....))))).((((((...(((........)))...))))))..........))))))............. (-21.95 = -22.52 + 0.57)

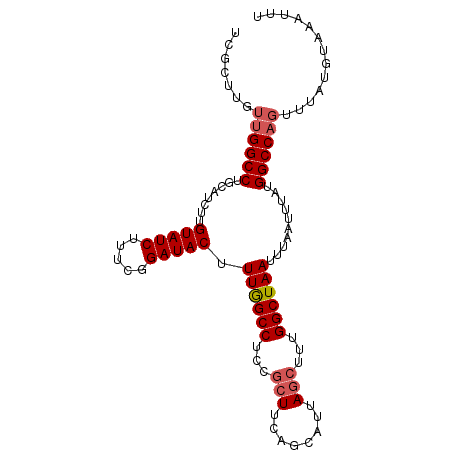
| Location | 11,229,718 – 11,229,816 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11229718 98 - 22407834 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAACGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.............(((...(((........)))...)))....((((.(....)))))........)))))........ ( -21.80) >DroPse_CAF1 119528 75 - 1 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCA--------------GCGACGGCUAAAGUAUCUAACAGAUACAAGAUGCAGGCC--------- ...............((((..........((((((.--------------.....)))))).(((((.....)))))........))))--------- ( -20.00) >DroSec_CAF1 61881 98 - 1 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.............(((...(((........)))...)))....((((.(....)))))........)))))........ ( -21.70) >DroSim_CAF1 65692 98 - 1 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.............(((...(((........)))...)))....((((.(....)))))........)))))........ ( -21.70) >DroEre_CAF1 62671 98 - 1 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCCGCAGCGAAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.........((((((....))))))...(((((....))....((((.(....)))))....))).)))))........ ( -25.60) >DroYak_CAF1 65270 98 - 1 AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCCGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.........((((((....))))))(((........)))....((((.(....)))))........)))))........ ( -22.90) >consensus AAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGCGA ..............(((((.............(((...(((........)))...)))....(((((.....)))))........)))))........ (-19.09 = -19.63 + 0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:40 2006